Journal of
eISSN: 2373-4345


Review Article Volume 10 Issue 4
Department of Oral Biology, Faculty of Fundamental and Biomedical Sciences, Paris Descartes University, France
Correspondence: Michel Goldberg, Professor Emeritus, Department of Oral Biology, Paris Descartes University, Faculty of Fundamental and Biomedical Sciences & INSERMUMR-S 1124. Stem cells, signalization and prions. 45 rue des saints pères, 75006, Paris, France, Tel 33162676709
Received: June 04, 2019 | Published: August 28, 2019
Citation: Goldberg M. Genetic and structural alterations of enamel and dentin- amelogenesis imperfecta, dentinogenesis imperfecta and dentin dysplasia. J Dent Health Oral Disord Ther. 2019;10(4):1?4. DOI: 10.15406/jdhodt.2019.10.00494
Genetic alterations of enamel and dentin include different sub-groups recognized on the basis of their clinical appearance. Ameloblasts secrete three major enamel ECM proteins: AMEL (amelogenin associated with Amelogenesis Imperfecta phenotypes, ranging from hypoplastic to hypomineralized enamel), AMBN (ameloblastin) and ENAM (enamelin) They are localized within a cluster of genes critical to biomineralization mapped on chromosome 4q21. Hypoplastic enamel displays secretory defects (pitted, rough or local). Hypomineralized (with eruption pathology), hypocalcified types with mineralization defects, and hypomature enamel result to altered protein processing and crystallite maturation defects. They display a chalky appearance, orange, brown or white colour. Enamel is pigmented, snow capped. Dentin defects are classified into three types of Dentinogenesis Imperfecta (DGI, types I-III) and two types Dentin Dysplasia (DDs, types I and II). DGI type II was originally called hereditary opalescent dentin or Capdepont’s teeth. Clinically, DGI-II is characterized by soft blue-brown, translucent teeth (opalescent teeth). Abnormal dentin obliterate the pulp chamber of DD type I. Genetically altered enamel and dentin structures allow significant insigths on dental tissue genetic alterations, and consequently increase our understanding of the formation of normal dental tissues. The affected dental tissues involve gene mutations, translated into structural proteins and/or implicated in the composition of dental tissues. This shed light on the cleavage of the constituent molecules of the ECM.
Keywords: proteins, molecules, phenotypes, dental tissues, gene mutations, heterogeneity, clinical appearance, autosomal recessive, basement membrane, maturation stage, enamel crystallites, gene
ECM, extracellular molecules; DD, dentin dysplasia; DI, dentinogenesis imperfecta; XLR, X-linked recessive; DSPP, dentin sialophosphoprotein; AR, autosomal recessive; AD, autosomal dominant
The protein gene family includes extracellular molecules (ECM) proteins, responsible for dentin/bone coding (DSPP, DMP1, IBSP, MEPE, and SPP1), enamel (AMEL, ENAM, AMBN, and AMTN), as well as milk casein, and some salivary protein genes (Table 1). These molecules encompass inherited defects of dental enamel (AI) and dentin (DI and DD). They display both clinical and genetic heterogeneity. These groups include different sub-types recognized on the basis of their clinical appearance. Diseases affecting tooth structures have been classified into distinct tissues [enamel (AI) versus dentin (DI & DD)], the specificity of the mutation (syndromic versus non-syndromic), and their pattern of inheritance [autosomal dominant (AD), autosomal recessive (AR), or X-linked recessive (XLR)]. Mutations in the AMELX , ENAM , MMP20 and KLK4 genes are associated with specific AI types. Another series of gene mutations influence dentin structure and composition [dentinogenesis imperfecta (DI) and dentin dysplasia (DD)]. These mutated genes are implicated in defective dental tissues.1–3
|
Gene symbol |
Protein name |
Protein distribution |
|
Ancestor |
|
|
|
SPARC |
secreted protein, acidic, cysteine rich (osteonectin) |
skeleton |
|
SPARCL1 |
secreted protein, acidic, cysteine-rich like 1 protein (high endothelial venule protein) |
brain |
|
SCPP |
|
|
|
DSPP |
dentin sialophosphoprotein |
dentin, bone |
|
DMP1 |
dentin matrix acidic phosphoprotein 1 |
dentin, bone |
|
IBSP |
integrin-binding sialoprotein (bone sialoprotein) |
dentin, bone |
|
MEPE |
matrix extracellular phosphoglycoprotein |
dentin, bone |
|
SPP1 |
secreted phosphoprotein 1 (osteopontin) |
dentin, bone |
|
AMEL |
amelogenin |
enamel |
|
ENAM |
enamel |
enamel |
|
AMBN |
ameloblastin (sheathlin, amelin) |
enamel |
|
AMTN |
(UNQ689) |
enamel |
|
ODAM |
odontogenic, ameloblast associated (APIN protein) |
milk, saliva, enamel |
|
FDCSP |
follicular dendritic cell secreted peptide |
milk, saliva, PDL |
|
MUC7 mucin 7 |
|
saliva |
|
PROL1 proline-rich 1 (basic proline-rich lacrimal protein 1) |
|
saliva |
|
PROL3 proline rich 3 [submaxillary gland androgen-regulated protein 3 |
|
|
|
homolog B (mouse)] |
|
saliva |
|
PROL5 (SMR3A) proline rich 5 [submaxillary gland androgen-regulated protein 3 |
|
saliva |
|
homolog A (mouse)] |
|
|
|
LOC401137 |
|
saliva |
|
HTN1 histatin 1 |
|
saliva |
|
HTN3 histatin 3 |
|
saliva |
|
STATH statherin |
|
saliva |
|
CSN3 |
k-casein |
milk |
|
CSN2 |
b-casein |
milk |
|
CSN1S1 |
aS1-casein |
milk |
|
PDL = Periodontal ligament; LOC401137 = the locus symbol given in the genome sequence database. |
|
|
|
Many salivary SCPPs are also present in tears. (reprinted from 4) |
|
|
Table 1 SCPP genes and ancestors
Amelogenin imperfecta
In mammals, ameloblasts secrete three major enamel ECM proteins: AMEL (amelogenin associated with AI phenotypes, ranging from hypoplastic to hypomineralized enamel), AMBN (ameloblastin) and ENAM (enamelin localized within a cluster of genes critical to biomineralization, mapped on chromosome 4q21). Mutations result in enamel hypoplasia. In addition, AMTN (amelotin) is preferentially expressed by ameloblasts, in the incisor and molar basement membranes, but exclusively during the late maturation stage.4,5
Mutations of MMP20 and KLK4 are proteinases critical for processing enamel matrix components. They are located on chromosomes 11q23 and 19q13 respectively.6, 7 In humans, enamel defects are including several types of AI, leading to enamel hyploplasia or hypomineralization. They are also known as hereditary enamel dysplasia, hereditary brown enamel, or hereditary brown opalescent teeth.8–12 AI is an heterogeneous group charcterized by defects in the formation of enamel due to mutations in AMELX (14 X-linked AI, AIH1), and/or in ENAM (5 autosomal-dominant AI, and AIH2 genes). More than 50 mutations have been identified, based upon the phenotypes and the mode of inheritance.
In enamel, three main groups have been reported: 8–12
X-linked alterations in the human amelogenin gene (AMELX) have been already reported.7
The classification of the IV types of AI was revisited.8,15,16 Now, this classification includes the following phenotypes:
|
Type I Hypoplastic |
|
Type II Hypomaturation |
|
Type III Hypocalcified |
|
Type IV: Hypomaturation-hypoplastic with taurodontism |
Mutation of the gene encoding MMP-20 (enamelysin) is located in the intron 6 splice acceptors. Enamel is pigmented, with a mottled rough surface.10 During development and mutations, the kallikrein 4 (KLK4) and enamelysin (MMP20) genes cause autosomal recessive AI.6, 7 Integrin, 6 is a member of a large family of cell-surface-adhesion receptors facilitating interactions with the cytoskeleton.14
Four enamelin gene (ENAM) defects have been identified in order to gain information related to genotype/phenotype correlations. The IVS6-2A<C mutation exhibits horizontal groves of hypoplastic enamel. In g.8344delG mutation, a generalized hypoplastic enamel is observed with shallow horizontal groves in the middle 1/3 of anterior teeth.8, 9
Laminin (LAMA3), and LAMB3 mutation are also at the origin of amelogenesis imperfecta,12 including the mutation in the last exon of LAMB3. Enamel formation is particularly sensitive to defects in hemi-desmosome/basement membrane complexes. The syndromic and non-syndromic forms of AI are etiologically related to this mutation.
Laminin (formerly laminin V) is a component of basement membranes and comprised 3 subunits. Laminin is anchored to epithelial cells by collagen XVII and activates integrin signaling through α6/β4 receptors.11–13 AMTN (Amelotin) is a proline, leucine, threonine and glutamine-rich protein, secreted during the transition and maturation stage of ameloblasts. The molecule bind to itself, to ODAM (odontogenic, ameloblast-associated) and to SCPPPQ1 (secretory calcium-binding phosphoprotein-glutamine-rich 1).5 COL17A1 is expressed throughout enamel formation.
Non-syndromic AI-causing mutations have been found in genes encoding secreted enamel matrix proteins and proteases (AMELX, ENAM, C4orf26, MMP20, KLK417-(12-14), intracellular (FAM83H, WDR72), transmembrane (SLC24A4, COL17A1), and basement membrane (LAMA3, LAMB3) proteins (Figure 2).15,16-19
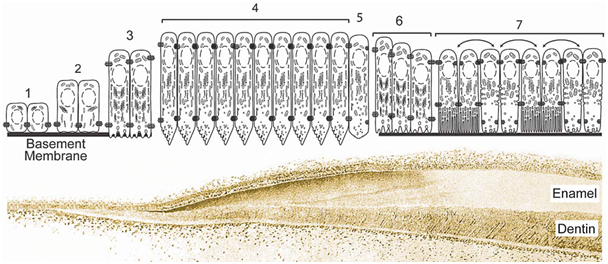
Figure 2 Reprinted from.13
Dentinogenesis imperfecta (DG1)17–21
Dentin defects are classified into three types of DI (DGI, types I-III) and two types DD (DDs, types I and II).20,21 In DGI, the dentin is defective and soft. In DD, the pulp chambers may be obliterated by abnormal dentin. Mutations of DSPP cause DD type II. They have been mapped to chromosome 4q21. The incidence of DI is evaluated between 1/6000 and 1/8000 children.
The known mutations of collagen genes and their phenotypic effects in OI is increasing, but so far, no definitive relation has been established between the type of OI and the manifestations of the dental defects.
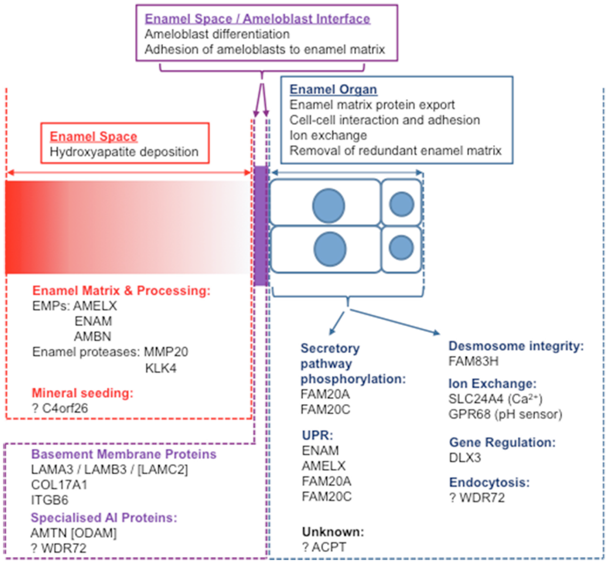
Figure 3 According to.19
Dentin dysplasia (DD): genes involved in dentin dysplasia formation
DD type I (DD-1) : Abnormal dentin obliterate the pulp chambers of DD type I. The prevalence of type I and II DD is about 1/100 000. They have short roots. Early exfoliation and periapical radiolucencies are noted. DD type II is detectable in deciduous teeth but not in permanent teeth. They have large pulpolithes (denticles or pulp stones) within the pulp. The SIBLING family of proteins includes dentin sialophosphoprotein (DSPP), osteopontin, IBSP (integrin binding sialoprotein), DMP-1 and MEPE (matrix extracellular phosphoglycoprotein). They display genes that are clustered on chromosome 4q. DSPP is located at 4q22.1 and consists of 5 exons. Three distinct protein products are formed from the initially translated polypeptide (DSPP): DSP, DGP and DPP. Dentin sialoprotein (DSP) and dentin phosphoprotein (DPP) overexpression are produced by the cleavage of a single gene (DSPP). After an initial cleavage producing the release of DPP, MMP20 generate DSP and DGP. Immediately after cleavage, DPP moves to the mineralization front and binds to collagen. DGP contains 4 phosphorylated serines and one N-glycosylated asparagine. The clinical phenotypes associated with DSPP mutations appear to represent a continuum of phenotypes. Thus, these disorders should be called DSPP-associated dentin defects, with DD type II representing the mild end of the phenotypic spectrum and DI type III expressing the critical end.
Dentin dysplasia type I (DD-1) is usually associated with osteogeniesis imperfecta. It is a consequence of the disintegration of Hertwig’s epithelial root sheath and the subsequent migration of epithelial cells to the dental papilla. It regulates the induction of synthesis of dentin matrix. The root formation is defective. The pulp chambers of deciduous teeth are completely obliterated, whereas a crescent-shaped pulpal remnant persists in the permanent teeth. Col1A1 and ColIA2 are encoded by the COL1 A1 and COL1 A2 loci at 17q21.3-q22 and 7q22.1, respectively.
Dentin dysplasia type II (DD-2) or hereditary opalescent dentin: the abnormal dentin matrix has been reported to stain positively for reticulin, suggesting the presence of type III collagen. Abnormalities of noncollagenous components include changes in the amount of different glycosaminoglycans and the presence of fibronectin. The deficiency in dentin phosphoprotein is common in types I and II DI. The pulp chambers of deciduous teeth are completely obliterated. In permanent teeth, denticles are found.20,21
To conclude, genetically altered enamel and dentin structures allow significant insigths on dental tissue abnormalities, and consequently on our understanding of the formation of normal dental tissues. The affected tissues (enamel, dentin and pulp) involve gene mutations, consequently translated into structural proteins and/or implicated in the composition of dental tissues.12,15–17 This enlighten also the cleavage of the constituent molecules of the ECM (Figure 4).
None.
None.
The authors declare that there is no conflict of interest.