Journal of
eISSN: 2373-633X


Research Article Volume 15 Issue 4
1Biochemistry Laboratory, Faculty of Medicine of Sousse, University of Sousse, Tunisia
2Biology Department, Hail University, Saudi Arabia
3Laboratory of Genetics, CHU Farhat Hached Sousse, Tunisia
4UR Cytogenetics, Molecular Genetics and Human Reproductive Biology, University of Sousse, Tunisia
5Biological Hematology Department, CHU Pontchaillou, France
6Hematology Biology Department, CHU A. Othmana, Tunisia
7Clinical Hematology Department, CHU F. Hached, Tunisia
Correspondence: Sawsen Besbes, Research unit “molecular biology of leukemia and lymphomas” (research unit 14es19), Bio-chemistry laboratory, Faculty of Medicine of Sousse Avenue Mohamed Karoui – 4000 Sousse, Tunisia
Received: July 25, 2024 | Published: August 9, 2024
Citation: Besbes S, Bouali N, Hamadou WS, et al. Molecular characterization of Tunisian B-acute lymphoblastic leukemia.J Cancer Prev Curr Res. 2024;15(4):77-80. DOI: 10.15406/jcpcr.2024.15.00555
B-Acute lymphoblastic leukemia (B-ALL) represents a heterogeneous spectrum of lymphoid disorders and stands as the most common hematological malignancy affecting both children and adults. The diagnosis generally based on morphological criteria as well immunophenotyping, while molecular approaches provide highly valuable clinical and prognostic information. In this study, our aim was to investigate IGH, IGK-Kde, and IKZF1 genes as molecular markers to enhance the accuracy of B-ALL diagnosis. Therefore we explored 63 B-ALL Tunisian cases, using multiplex PCR assay according to BIOMED-2 condition. 34 clonal IGH gene rearrangements, 22 clonal IGK-Kde, 4 IKZF1 gene deletions and 2 simultaneous IG/IKZF1 recombination were identified. These findings confirm both the clonal proliferation and the B-lymphoid lineage origin. The use of IGH, IGK-Kde and IKZF1 markers will be introduced for the first time in Tunisian laboratories for molecular characterization of B-ALL and subsequently for the monitoring of minimal residual disease which is an important determinant for patients outcome. The implementation of molecular profiling of B-ALL trough assessing IGH, IGK-Kde, and IKZF1 markers will rise the challenge for efficient minimal residual disease monitoring and patient outcomes evaluation.
Keywords: B-ALL, IGH rearrangement, IGK rearrangement, IKZF1 deletion, multiplex PCR
Acute lymphoblastic leukemia (ALL) is a malignant disorder originating from lymphoid progenitor cells committed to differentiating into T or B cells. Its origin is attributed to significant genetic abnormalities in precursor blood cells. ALL is the most common hematological malignancy with B-cell origin (B-ALL) with predominance ranging from 75% to 92%.1 Assessing clonality through targeted molecular markers can significantly support the diagnosis of B-ALL. Studies highlight that 5 to 15% of leukemia cases could benefit from molecular clonality assessment.2 Unlike cytomorphology, molecular biology approach aligned with immunophenotyping, allow best characterization of leukemic blasts by confirming the monoclonality and the B-lineage origin of the malignant proliferation. Several molecular methods have been developed to identify leukemic population. The immunoglobulin (IG) gene rearrangements are the most widely applied targets. Heavy chain (IGH) and light chain (IGK/IGL) genes recombination are commonly used for B-leukemia characterization.3,4 During the IGH locus rearrangement, the VHDHJH recombination involves one of the VH gene from the seven subgroups (IGHV1-IGHV7). The similarity in rearrangements among these subgroup members primarily arises from their high sequence homology. This similarity enables the amplification of the IGH gene using consensus primers via molecular methods.5–7
For IGL locus rearrangement, the IGK locus is deleted involving the kappa deleting element (Kde). Two different types of Kde recombination can inactivate the IGK rearrangements: Kde rearranges to the intron RSS leading to the deletion of the IGKC gene and the maintain of the IGKV-J junction, or Kde rearranges to one of the VK genes leading to the exclusion of constant region and intron RSS with the subsequent loss of the IGKV-J junction.8,9 Therefore, it is possible to detect both IGKV-J and intron-Kde deletional rearrangements on the same allele.8,9
During each IGH or IGK recombination event, nucleotides are inserted at the junctions of VDJ or VK-Kde/RSS-Kde, respectively. This process creates N-regions, which serve as unique molecular sequence tags specific to each lymphocyte. These tags are crucial for identifying and characterizing the diversity of lymphocytes based on their specific genetic rearrangements.10,11 Since almost 90% of B-ALL rearrange the IGH locus and 50% delete the IGK-Kde locus, these molecular markers are commonly used for of B-leukemia diagnosis.
Similarly, the Ikaros protein mediates chromatin accessibility necessary for VDJ recombination add to modulates the expression of early B-cell-specific genes.12 Ikaros is encoded by Ikaros Family Zinc Finger 1 gene (IKZF1) and contains variable number of Kruppel-like zinc fingers in two domains involved in DNA binding.13 IKZF1 deletions are closely related to the development of lymphoid leukemia and found in nearly 30% B-ALL Ph- .11 The most common deletions (~45%) occur in exons 4 to 7. The 5’ and 3’ breakpoints are highly conserved and are flanked by recombination signal sequences RSS. This suggests that this deletion is mediated by off-target effects of the recombination activating gene during VDJ recombination in B cell progenitors.14,15 The deletions Δ4-7 and Δ2-7 are the most frequently identified. IKZF1 deletions are associated with a poor prognosis in pediatric and adult B-ALL and can be an efficient predictor of relapse.16
Patient risk stratification may be improved by comparing IKZF1 status to other adverse prognosis criteria.16 The B-ALL diagnosis is based on a combination of clinical assessment, cytological examination, cytogenetic analysis, and immunophenotypic classification. These approaches may often present some difficulties of interpretation, therefore, more sensitive and complementary methods are requested for leukemia diagnosis and patients’ treatment stratification. We suggest utilizing a combination of molecular markers, such as IKZF1 deletions and IGH/IGK-Kde rearrangements, to enhance the diagnosis of B-ALL in Tunisian patients."
Patients
63 Tunisian B-ALL cases were included in this study. 44 B-ALL were diagnosed at Hematology department, F. Hached Hospital of Sousse and 19 diagnosed at Biological Hematology department, A. Othmana Hospital of Tunis. Informed consent was obtained from all patients or their legal guardian as required by the Helsinki Declaration. The cohort of patients consisted of 38 males and 25 females with sex ratio: 1,52 and median age of 8 years at the time of diagnosis (1-57 years). The B-ALL were characterized based on the French-American-British classification and flow cytometric immunophenotyping.
Mono nuclear cells and DNA isolation
Samples were collected from either bone marrow (23 patients) or peripheral blood (40 patients) upon diagnosis, where blast count exceeded 75%, ensuring detectable analysis of IGH and IGK-Kde rearrangements, as well as IKZF1 deletions. Mononuclear cells were isolated using Ficoll-Paque density centrifugation and were subsequently utilized for DNA extraction. Genomic DNA extraction was performed using either phenol chloroform or the QIAGEN DNA Blood kit as per the manufacturer’s protocols. DNA quantity was determined using spectrophotometry, and its quality was assessed via agarose gel electrophoresis.
PCR analysis
Clonal IGH and IGK-Kde gene rearrangements and IKZF1 deletion were investigated by multiplex PCR amplification. For IGH gene rearrangements, VH family specific primers combined with a consensus JH primer were used.17 For IGK-Kde rearrangements, a reverse Kde primer was used in combination with one of six VK family-specific primers or with RSS primer located at the JK-CK intron.17 For IKZF1 deletion, specific Δ2-7, Δ2-8, Δ4-7 and Δ4-8 primers we used as previously described (primers are available under request).16 PCR amplifications were conducted in triplicate following the BIOMED-2 consortium protocol.16 PCR products were subsequently identified using Gene Scanning on the ABI Prism 3130xl Genetic Analyzer (Applied Biosystems).
Clinical and biological characteristics
The hyperleukocytosis observed in 63 patients in favor of B-ALL clinical suspicion that reflects the high tumor burden. Cytomorphological examination highlighted transient massive blastosis in peripheral blood, along with bone marrow infiltration by leukemic cells and extramedullary manifestations. The myelogram revealed more than 20% blasts, with immunophenotyping indicating low CD45 intensity, allowing gating of blasts expressing HLA-DR and CD34. B-ALL cases were positive for CD79a, CD22, and CD19, classified predominantly as B-ALL III according to EGIL classification.
Molecular characteristics
The validity of the extracted leukemic DNA was confirmed by RQ-PCR, as the obtained threshold cycle (Ct) differed by no more than ±1 Ct from the control DNA (with Ct values of 23.9 for 50ng and 20.5 for 500ng of DNA). Only 55 genomic DNAs among 63 were investigated for IGH / IGK-Kde and IKZF1 loci analysis due inadequate DNA quality. The multiplex PCR was performed using standard primers specific to either IGH or IGK-Kde loci. 56 clonal IGH and IGK-Kde rearrangements were revealed. For IGH rearrangement assessment, using the combination of 7 primers (VH1 to VH7) via multiplex PCR assay, 34 B-ALL VH-JH rearrangement were revealed with one pic at the expected size 250-295 pb (Figure 1). The majority of rearrangements included VH3, VH4 genes; followed by VH1, VH2, VH5 and VH7. For IGK-Kde rearrangement, a combination of 7 primers (VK1f/6 to VK7 and intron RSS) via multiplex PCR assay; In 22 B-ALL, we obtained one pic at the expected size of 120-300 pb corresponding to VK-Kde or RSS-Kde rearrangement (Figure 2). The VK1f/6-Kde rearrangement was predominant one. The recombination events involving Kde were found to include VK2, VK3, VK7, and the intron RSS sequences. Four B-ALL cases showing bi-allelic IGK-Kde rearrangements: two cases revealed VK1f/6-Kde on the first allele and VK7-Kde on the second one, and two cases exhibited VK3-Kde on one chromosome and intron RSS-Kde on the second allele. Concomitant rearrangements of both IGH and IGK loci were observed in 14 B-ALL cases. The investigation of IKZF1 gene was performed in 39 cases by targeting Δ2-7, Δ2-8, Δ4-7 and Δ4-8 deletions using multiplex PCR assay. We obtained one pic at the expected size of 120-195 pb corresponding to the Δ4-7 deletion identified in 3 B-ALL and one pic at the expected size of 250-280 pb corresponding to the Δ2-8 deletion (Figure 3).
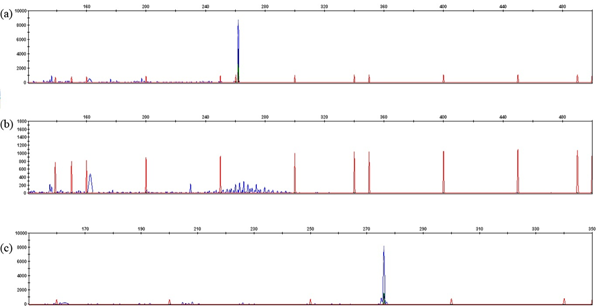
Figure 1 Gene scan of IGH rearrangements: a) monoclonal control DNA, b) polyclonal control DNA, c) monoclonal IGVH1(P14).
B-ALL represents the predominant subgroup among hematological malignancies, affecting both children and adults (comprising 60% to 80% of all cases of ALL). For a comprehensive characterization of leukemic lymphoblasts, it is essential to integrate cytological diagnosis and immunophenotyping with molecular approaches. This ensures the most accurate classification and understanding of the disease. Through our study, we introduced the molecular diagnosis approaches for accurate diagnosis and monitoring of B lineage leukemias, since this method is not commonly used in laboratories. Among standard molecular markers, PCR-based analysis of immunoglobulin rearrangements has gradually become a gold standard method. IGH, IGK-Kde rearrangements can be commonly used for B-ALL diagnosis since they are identified in nearly 90% and 50% of cases respectively.18,19 Moreover, IKZF1 is deleted in 28% of B-ALL and shows poor prognostic mainly in B-ALL Ph-.20 Therefore, we combined IGH / IGK-Kde rearrangements and IKZF1 deletions as molecular markers for the first time to assess Tunisian B lineage leukemia.
We have analyzed 63 B-ALL cases based on cytomorphological criteria and immunophenotyping. The IGH and IGK-Kde multiplex PCR assays were performed using BIOMED-2 conditions. The presence of clonal proliferation was confirmed in 34 B-ALL through IGH rearrangements. For VHJH rearrangement, the VH gene involvement was similar to previous reported studies. The VH3 and VH4 were the most commonly genes involved (26% vs 20%-50%), followed by VH1 (17% vs 10%-20%), VH2 (6% vs 5%-11%) and VH5 (3% vs 3%-4%).21 The VH6 gene rearrangement was not detected in our investigated cases despite it is frequently rearranged in B-ALL according literature.17 We have demonstrated the IGK gene deletions in 22 cases of B-ALL, characterized by Kde segment rearrangements predominantly involving the Vκ1f/6 gene (50% vs 68%-100%), and less frequently with intron RSS (14% vs 0-31%).3s,17,22
In our cohort, 64% of B-ALL involved concomitant IGH and IGK-Kde rearrangements. These two combined genetic markers are therefore two powerful and helpful marker for the clonality assessment and the accurate diagnosis for the vast major of B-ALL subtypes. Using multiplex PCR, the deletion of IKZF1 gene were detected in 10% of B-ALL investigated cases compared to 9%-20% as described in literature.23 Therefore, we have verified that the lymphoblastic proliferation is clonal, with the Δ4-7 deletion in IKZF1 identified as the most frequent deletion. Most IKZF1 intragenic deletions including exons 4 to 7, drive the expression of a non-DNA binding Ikaros isoform with dominant negative activity. This implies an active, ongoing recombination at IKZF1 deletion junction during the leukemogenic process.23,24 The significant correlation between IKZF1 mutations and poor prognosis has led to recommendations for screening IKZF1 mutations upon diagnosis.16 In children, IKZF1 deletions are a better predictor of relapse compared to conventional classification based on clinical and cytogenetic criteria.24 This suggests that risk stratification of patients could be enhanced by comparing IKZF1 status with other adverse prognosis criteria, such as residual disease status. Interestingly, in our study, two B-ALL cases exhibited simultaneous modifications in the IGH, IGK-Kde, and IKZF1 genes. These findings strongly support the diagnosis of clonality and suggest potential for improved patient stratification to enhance follow-up and refine therapeutic protocols.
Here, we have employed a molecular approach to successfully detect combined IGH/IGK-Kde rearrangements and IKZF1 deletions for diagnosing B-ALL patients. We aim imply this approach in Tunisian laboratories. The tags of both IG and IKZF1 would be a helpful markers for best characterization of B-ALL to complete cytomorphology and immunophenotyping approaches. The loss of IKZF1 may interact with IGH rearrangements and other biological or clinical characteristics to define subsets of leukemia that are particularly aggressive. When IKZF1 mutations are detected at diagnosis, they may be reliable markers to follow minimal residual disease (MRD).16,23 We will further introduce both IG and IKZF1 genes to follow-up B-ALL patients during treatments and assess the MRD which is powerful prognosis indicator.
This study was approved by the ethics committee of the hospital and all subjects gave their informed consent according to Helsinki declaration.
This work was supported by the Ministère de l’Enseignement Supérieur et de la Recherche Scientifique en Tunisie.
The author declares that there are no conflicts of interest.

©2024 Besbes, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.