Journal of
eISSN: 2469 - 2786


Research Article Volume 13 Issue 1
1Microbiology Department, Department of Biotechnology Research, Ministry of Science and Technology, Myanmar
2Department of Plant Medicine, Kyungpook National University, South Korea
Correspondence: San San Yu, Professor, Microbiology Department, Department of Biotechnology Research, Kyaukse, Myanmar
Received: March 01, 2025 | Published: March 24, 2025
Citation: San SY, Hee-Young J. Isolation and molecular identification of Parasediminibacterium sp. BT-541 isolated from soil. J Bacteriol Mycol Open Access. 2025;13(1):52-56. DOI: 10.15406/jbmoa.2025.13.00398
A Gram-negative, orange-colored bacteria, designated BT-541, was isolated from a soil sample in South Korea. Comparative analysis of 16S rRNA gene sequences revealed that the strain BT-541 exhibited the highest sequence similarity with Parasediminibacterium paludis HME6815T (98.50%) and 94.2-93.12% sequence similarity with other members of the family Chitinophagaceae. This strain could grow at temperatures ranging from 15 °C to 37 °C and pH ranging from 6-10. The only respiratory quinone in the bacterial isolate was menaquinone 7 (MK-7), and the major fatty acids were iso-C15:0, iso-C15:1G, and iso-C17:0 3-OH. Polar lipids included a phosphatidylethanolamine, three unidentified aminolipids, three unidentified aminophospholipids, an unidentified phospholipid, and five unidentified lipids. Molecular analysis and phenotypic characteristics indicated that strain BT-541 (KCTC 82123 = JCM 37057 = PQ 898028) could be a potential novel species in the genus Parasediminibacterium.
Keywords: Parasediminibacterium, Chitinophagaceae, 16S rRNA, respiratory quinone, fatty acid, polar lipids
The bacterial genus Parasediminibacterium, classified under the phylum Bacteriodetes, class Chitinophagia, order Chitinophagales, and family Chitinophagaceae, was initially introduced through the identification of Parasediminibacterium paludis by researchers Heeyoung Kang and colleagues.1 Subsequently, Kämpfer and his team documented the Chitinophagaceae family,2 highlighting its significant phylogenetic diversity within the Bacteriodetes phylum.3 Currently, the Chitinophagaceae family consists of 51 recognized genera, a number recently expanded by the discovery and naming of Foetidibacter,4 Ginsengibacter,5 Phnomibacter,6 Pseudocnuella,7 Agriterribacter,8 Mucibacter,9 Parapseudoflavitalea,10 and Paraflavitalea.11 Kämpfer and Kim noted that bacteria belonging to the Chitinophagaceae family are typically aerobic or can survive in both aerobic and anaerobic environments, usually lack motility, exhibit a rod-like shape, and primarily utilize menaquinone-7 (MK-7) as the isoprenoid quinone, along with iso-C15:0, iso-C15:1 G, and iso-C17:0 3-OH as the major fatty acids.12,13 To date, the genus Parasediminibacterium consists of a single genus and species (https//lpsn.dsmz.de/genus/Parasediminibacterium). Therefore, more research is required to determine the functions of the Parasediminibacterium strain. The discovery of a new Parasediminibacterium species adds to the genus' variety and bolsters theoretical frameworks like genetic and genomic research for more relevant studies. This study isolated bacterium BT-541 and examined its phylogenetic, physiological, and biochemical characteristics. Despite being a member of the genus Parasediminibacterium, the strain has a number of physiological and biochemical traits that set it apart from Parasediminibacterium paludis. As a result, this strain may represent a new species of Parasediminibacterium. Investigating potential novel species of native bacteria isolated from soil samples in South Korea was the goal of this study in order to improve our understanding of their diversity, ecology, and distribution for potential use in a variety of industries, and agricultural sector.
Sample collection and strain isolation
A novel orange-colored bacterial strain BT-541, was isolated from a soil sample in South Korea. One gram of soil sample was suspended in ten milliliters of 0.85% saline solution and shaken for half an hour. After setting for 2 hours, the sample solution was serially diluted (10−1 to 10−4). Then, 100 μl of the serially diluted soil solution was spread out over Reasoner's 2A (R2A) agar (Difco, Detroit, MI, USA) and incubated at 30 °C for seven days.14 On R2A medium, one orange colony was selected and subcultured. The isolate was then purified by streaking it on the same medium for several times. For further study, the pure culture of strain BT-541 was regularly maintained at -80 °C in 20% (v/v) glycerol after being regularly cultivated on Reasoner's 2A (R2A) agar at 30 °C. The strain BT-541 has been deposited to the Japan Collection of Microorganisms (JCM 37057), the Korean Collection for Type Cultures (KCTC 82123), and the NCBI GenBank accession number (PQ 898028).
Morphological and physiological analysis
The growth temperature range of strain BT-541 was assessed on R2A agar at 4, 10, 15, 20, 25, 30, 37, 42, and 45 °C. The pH range of the growth was evaluated at pH 4.0–10.0 (at 1.0 pH unit intervals) on R2A agar after 7 days of incubation. The pH of the medium was adjusted with 1M NaOH and HCl. Growth in the presence of 0.5 % and 1.0–5.0 % (at intervals of 1.0 %) NaCl (w/v) was tested on R2A agar after 7 days. Growth of the isolated strain was observed on nutrient (NA; Difco), marine 2216 (MA; Difco), tryptic soy (TSA; Difco), Luria Bertani (LB; Difco), and MacConkey (Difco) agars. In accordance with the manufacturer's instructions (bioMérieux), additional biochemical tests were conducted using API ZYM, API 20NE kits, and API ID 32 GN microtest systems, and the tested bacterial isolate's results were documented.
Phylogenetic analysis
The The HiGene Genomic DNA Prep Kit (Biofact, Daejeon, Korea) was used to extract the whole genomic DNA of BT-541 from cultures that were cultivated on R2A for 72 hours. The approach outlined by Weisburg et al.16 served as the foundation for the polymerase chain reaction (PCR) protocol utilized in this investigation to amplify the 16S rRNA region. 9F (5'-GAG TTT GAT CCT GGC TCA G-3') and 1512R (5'-ACG GCT ACC TTG TTA CGA CTT-3') were the primers utilized in this investigation [16]. ExoSAP-IT PCR Product Cleaning Reagent (Thermo Fisher Scientific, Waltham, MA, USA) was used to purify the amplified PCR product of strain BT 541. SeqMan software (DNASTAR, Madison, WI, USA) was used to construct the whole 16S rRNA gene sequence. In order to guarantee the quality and comprehensiveness of the gene sequences, BioEdit version 5.0.6 was utilized to manually edit any gaps and terminal ends of the alignment. The EzBioCloud server and BLAST searches based on the 16S rRNA gene were used to identify phylogenetically closest sequences and calculate pairwise sequence similarity values. The CLUSTALW tool was used to construct multiple alignments of the 16S rRNA gene sequences of related species that were retrieved from GenBank.17 The MEGA 7 program's neighbor-joining approach was used to create a phylogenetic tree.18 The two-parameter approach developed by Kimura was used to calculate evolutionary distances. Bootstrap analysis with 1,000 repeats was used to assess the robustness and dependability of the resulting clusters.
Chemotaxonomic analysis
When the cells reached the mid-exponential development phase on R2A agar at 30 °C, the cellular fatty acids of strain BT-541 were extracted, saponified, and methylated in accordance with the standard methodology of MIDI (Sherlock Microbial Identification System). Gas chromatography was used to evaluate the fatty acids, and the Microbial Identification Software (TSBA, version 6.0) was used to identify them.19 According to Minnikin et al.,20 polar lipids and isoprenoid quinones of strain BT-541 were isolated and examined from freeze-dried cells. Two-dimensional TLC (silica-gel-coated plates, 10 cm × 10 cm; Merck) was used to investigate lipid separation. In the first dimension, the solvents were chloroform/methanol/water (65:25:4, v/v), while in the second dimension, the solvents were chloroform/acetic acid/methanol/water (80:15:12:4, v/v). The plates were sprayed with molybdenum blue (Sigma) for phospholipids, ninhydrin (Sigma) for aminolipids, α-naphthol for glycolipids, and 10% ethanolic molybdophosphoric acid (Sigma) for total polar lipids.
Morphological and physiological analysis
The cells of strain BT-541 are Gram-stain-negative, typically measuring between 1-5 and 3 µm in length and 0.4-0.5 µm in diameter, and are rod-shaped bacteria (Table 1 &2). Observations showed good growth on R2A agar and Nutrient agar, while no growth was detected on Marine 2216, Tryptic Soy, Luria Bertani, and MacConkey agars. Growth occurred on 1/10 Tryptic Soy agar. Strain BT-541 was capable of growing within a pH range of 6-10 and a temperature range of 15-37 °C, with the optimal conditions being a pH of 7 and a temperature of 30 °C. Several traits differed from those of the closely related strain, Parasediminibacterium paludis. Additionally, it displayed distinct characteristics compared to other reference strains in terms of β-glucosidase activity and glycogen assimilation. The detailed morphological, physiological, and biochemical characteristics of strain BT-541 are given in the species description and in Table 3.
|
Species |
Strain |
GenBank accession numbers |
|
Asinibacterium lactis |
LCJ02T |
JQ638910 |
|
Ferruginibacter alkalilentus |
HU1-GD23T |
FJ177530 |
|
Hydrotalea flava |
CCUG 51397T |
NR117026 |
|
Hydrobacter penzbergensis |
EM 4T |
JQ717375 |
|
Hydrotalea sandarakina |
AF-51T |
JF739858 |
|
Parasediminibacterium paludis |
HME6815T |
HQ231219 |
|
Parasediminibacterium sp. |
BT-541 |
PQ898028 |
|
Sediminibacterium aquarii |
AA5T |
KR812546 |
|
Sediminibacterium ginsengisoli |
DCY13T |
EF067860 |
|
Sediminibacterium goheungense |
HME7863T |
JN674641 |
|
Sediminibacterium magnilacihabitans |
MU-2T |
FJ816610 |
|
Sediminibacterium roseum |
SYL 130T |
KY922827 |
|
Sediminibacterium salmoneum |
NJ-44T |
EF407879 |
|
Sediminibacterium soli |
WSJ-3T |
MT299774 |
Table 1 1 List of type strains used for phylogenetic analysis and GenBank accession numbers of their 16S rRNA gene sequences
The isolated strain is shown in bold.
|
Species |
Strain |
Other collection |
Isolation source |
Year of isolation |
Geographic origin |
Reference |
|
Parasediminibacterium sp. |
BT-541 |
KCTC82123 |
Soil |
2013 |
South Korea |
This study |
|
JCM37057 |
||||||
|
PQ898028 |
||||||
|
Parasediminibacterium paludis |
HME6815T |
KCTC23736T |
Water |
2016 |
South Korea |
1 |
|
CECT8010T |
||||||
|
Sediminibacterium salmoneum |
NJ-44T |
CGMCC1.6845T |
Sediment |
2008 |
P. R. China |
21 |
|
NBRC103935T |
||||||
|
Sediminibacterium ginsengisoli |
DCY13T |
KCTC12833T |
Soil |
2013 |
South Korea |
22 |
|
JCM15794T |
||||||
|
DSM22335T |
||||||
|
Sediminibacterium goheungense |
HME7863T |
KCTC23945T |
Water |
2014 |
South Korea |
23 |
|
CECT8100T |
Table 2 The isolate BT-541 and reference strains used in this study
|
Characteristic |
1 |
2 |
3 |
4 |
5 |
|
Growth at |
|
|
|
|
|
|
20 °C |
+ |
- |
+ |
+ |
+ |
|
37 °C |
+ |
- |
+ |
+ |
+ |
|
Degradation of gelatin (API 20NE) |
+ |
- |
- |
- |
+ |
|
Enzyme activities (API ZYM) |
|
||||
|
Esterase (C4) |
w |
- |
- |
- |
- |
|
Esterase lipase (C8) |
w |
- |
- |
- |
- |
|
Cystine arylamidase |
w |
+ |
+ |
- |
+ |
|
Trypsin |
+ |
+ |
+ |
+ |
+ |
|
α-Chymotrypsin |
+ |
- |
- |
+ |
+ |
|
α-Galactosidase |
w |
- |
- |
+ |
- |
|
β-Galactosidase |
+ |
- |
+ |
- |
- |
|
β-Glucuronidase |
- |
- |
- |
- |
- |
|
α-Glucosidase |
+ |
+ |
+ |
- |
+ |
|
β-Glucosidase |
+ |
- |
- |
- |
- |
|
α-Mannosidase |
- |
- |
- |
- |
- |
|
α-Fucosidase |
w |
- |
- |
- |
- |
|
Assimilation of (API 32 GN) |
|
||||
|
Sucrose |
+ |
- |
- |
w |
+ |
|
Maltose |
+ |
- |
- |
+ |
+ |
|
D-Glucose |
+ |
- |
- |
+ |
+ |
|
Melibiose |
+ |
- |
- |
+ |
+ |
|
D-Mannitol |
- |
ND |
+ |
- |
- |
|
L-Proline |
+ |
- |
- |
+ |
+ |
|
L-Alanine |
+ |
- |
- |
+ |
+ |
|
Glycogen |
+ |
- |
- |
- |
- |
|
DNA G+C content (mol %) |
|
38.4 |
43.4 |
47.5 |
40.5 |
Table 3 Differential characteristics of strain BT-541 and type strains of related species
Strains: 1, BT-541 (data from this study); 2, Parasediminibacterium paludis HME 6815T (data from Kang H et al., 2016);1 3, Sediminibacterium salmoneum KACC 15020T (data from Qu JH et al., 2008);21 4, Sediminibacterium ginsengisoli KCTC 12833T (data from Kim YJ et al., 2013);22 5, Sediminibacterium goheungense HME 7863T (data from Kang H et al., 2014).23
+, positive; -, negative; w, weak positive; ND, not determined.
Phylogenetic analysis
The 16S rRNA region of strain BT-541 has a sequence length of 1,400 bp. The 16S rRNA gene sequence of BT-541 exhibited the highest similarity to Parasediminibacterium paludis HME6815T at 98.50%, while it showed lower sequence similarities to other members of the Chitophagaceae family, including Sediminibacterium ginsengisoli DCY13T (94.20%), Sediminibacterium salmoneum NJ-44T (94.03%), Sediminibacterium goheungense HME7863T (93.88%), Sediminibacterium soli WSJ-3T (93.16%), Sediminibacterium roseum SYL130T (93.12%), and Sediminibacterium aquaria AA-5T (93.12%). As illustrated in Figure 1, a phylogenetic tree constructed using the Neighbor-Joining method revealed that strain BT-541 clustered in the same branch as Parasediminibacterium paludis HME6815T. Similar topologies were also observed in both the Maximum Likelihood and Maximum Parsimony trees.
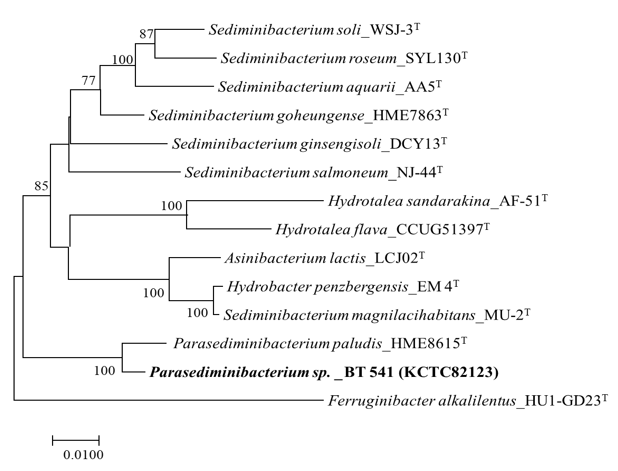
Figure 1 Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences, showing the phylogenetic positions of strain BT-541 and related taxa of the family Chitinophagaceae. Bootstrap support values of >70% are indicated at the nodes. The isolated strain is shown in bold. Ferruginibacter alkalilentus HU1-GD23T was used as an out-group. Bar, 0.0100 substitutions per nucleotide position.
Chemotaxonomic analysis
The fatty acid profiles of strain BT-541 and the four reference strains are outlined in Table 4. The predominant fatty acids found in strain BT-541 were C15:0 iso (20.13%), C15:1 iso G (18.32%), and C17:0 iso 3OH (9.91%). As indicated in Table 4, the overall fatty acid composition of strain BT-541 was quite similar to that of the three reference strains,21-23 with C15:0 iso, C15:1 iso G, and C17:0 iso 3OH being the major fatty acids; however, a higher concentration of C15:1 iso G and a lower concentration of C13:0 iso compared to the strain Parasediminibaterium paludis HME6815T were noted. Additionally, notable variations in the proportions of several components and the detection of a trace amount (<1%) of C17:0, C18:0, C17:1 ω5c, C18:1 ω9c, C11:0 iso, C14:0 iso 3OH, C13:1, summed feature 2 (C14:0 3OH/C16:1 iso I) and 7 (C19:1 ω7c/C19:1 ω6c/C19:0 cyclo ω10c/C19:0 ω6c) distinctly set strain BT-541 apart from other related strains. The polar lipids of strain BT-541 included a phosphatidylethanolamine, three unidentified aminolipids, three unidentified aminophospholipids, an unidentified phospholipid, and five unidentified lipids. Strain BT-541 was differentiated from the closely related strain Parasediminibaterium paludis HME6815 by the presence of an unidentified phospholipid. The sole isoprenoid quinone identified in strain BT-541 was menaquinone with seven isoprene units (MK-7), consistent with other members of the Chitinophagaceae family. Based on phenotypic and phylogenetic assessments, strain BT-541 may represent a new species within the genus Parasedimini bacterium.
|
Fatty acid |
1 |
2 |
3 |
4 |
5 |
|
Saturated |
|
|
|
|
|
|
C12:0 |
TR |
- |
- |
- |
TR |
|
C14:0 |
1.23 |
TR |
1.6 |
- |
TR |
|
C16:0 |
7.76 |
5.8 |
1.2 |
TR |
2.7 |
|
C17:0 |
TR |
- |
- |
- |
- |
|
C18:0 |
TR |
- |
- |
- |
- |
|
Unsaturated |
|
|
|
|
|
|
C15:1 ω6c |
3.62 |
3.7 |
1.1 |
- |
- |
|
C16:1 ω5c |
1.10 |
- |
TR |
- |
- |
|
C17:1 ω5c |
TR |
- |
- |
- |
- |
|
C17:1 ω6c |
1.57 |
2.7 |
TR |
- |
- |
|
C17:1 ω8c |
TR |
1.5 |
- |
- |
- |
|
C18:1 ω9c |
TR |
- |
- |
- |
- |
|
Branched -chain fatty acid |
|
|
|
|
|
|
C11:0 iso |
TR |
- |
- |
- |
- |
|
C13:0 iso |
3.96 |
11.0 |
4.3 |
2.9 |
4.1 |
|
C14:0 iso 3OH |
TR |
- |
- |
- |
- |
|
C15:0 iso |
20.13 |
14.3 |
17.5 |
21.8 |
20.3 |
|
C15:0 iso 3OH |
3.96 |
4.1 |
3.6 |
6.3 |
3 |
|
C15:1 iso G |
18.52 |
1 |
24.1 |
19.4 |
25.3 |
|
C16:0 iso |
1.13 |
5.7 |
1 |
- |
1.6 |
|
C16:0 iso 3OH |
2.73 |
7.9 |
2.6 |
3.4 |
6.6 |
|
C16:1 iso H |
TR |
1.7 |
- |
- |
- |
|
C17:0 iso |
TR |
TR |
- |
TR |
- |
|
C17:0 iso 3OH |
9.91 |
12.7 |
10.7 |
28 |
9.6 |
|
C13:0 anteiso |
TR |
- |
- |
- |
- |
|
C15:0 anteiso |
2.82 |
1.6 |
9.5 |
2.4 |
2.7 |
|
C15:1 anteiso A |
TR |
- |
7.4 |
- |
1.6 |
|
Hydroxy fatty acids |
|
|
|
|
|
|
C15:0 2OH |
TR |
1.2 |
TR |
TR |
1.6 |
|
C16:0 3OH |
6.46 |
2.8 |
1.8 |
3.3 |
2.4 |
|
C17:0 2OH |
2.02 |
2 |
2.2 |
1.6 |
TR |
|
C17:0 3OH |
TR |
TR |
- |
- |
2.1 |
|
Other |
|
|
|
|
|
|
C13:1 at 12-13 |
1.17 |
- |
- |
- |
- |
|
Summed feature* |
|
|
|
|
|
|
2; C14:0 3OH/C16:1 iso I |
TR |
- |
- |
- |
- |
|
3; C16:1 ω7c/C16:1 ω6c |
6.48 |
12.2 |
TR |
8.2 |
6.8 |
|
7; C19:1 ω7c/C19:1 ω6c/C19:0 cyclo ω10c/C19:0 ω6c |
TR |
- |
- |
- |
- |
|
9; C17:1 iso ω6c/C16:0 10-methyl |
TR |
1.7 |
- |
- |
- |
Table 4 Cellular fatty acid compositions of strain BT-541 and type strains of related species
Strains: 1, BT-541; 2, Parasediminibacterium paludis HME 6815T (data from Kang H et al., 2016);1 3, Sediminibacterium salmoneum KACC 15020T (data from Qu JH et al., 2008);21 4, Sediminibacterium ginsengisoli KCTC 12833T (data from Kim YJ et al., 2013);22 5, Sediminibacterium goheungense HME 7863T (data from Kang H et al., 2014).23
TR, Trace (<1.0%); -, not detected.
Values are percentages of total fatty acids.
*Summed Features are fatty acids that cannot be resolved reliably from other fatty acids using the chromatographic conditions chosen. The MIDI system groups these fatty acids together as one feature with a single percentage of the total.
The phylogenetic position of the strain BT-541 is in the same group with Parasediminibacterium paludis HME6815T, however, the specific association of phenotypic and chemotaxonomic features makes strain BT-541 clearly distinct from the already validly described Parasediminibacterium paludis HME6815T. Further research is needed to confirm the novel species and to reveal the ecological distribution of the strain BT-541.
Description of Parasedimini bacterium sp. BT-541
Cells are approximately 1-5 to 3 um in length and 0.4-0.5 um in diameter. Colonies of strain BT-541 on R2A agar are orange, smooth, convex, circular with entire margins, and approximately 1 mm in diameter after 3 days of incubation at 30 °C. Growth occurs at 10-37 °C (optimum 30 °C), at pH 6-10 (optimum pH 7.0), and cannot tolerate NaCl. Growth occurs on R2A agar, nutrient agar, and 1/10 tryptic soy agar. Growth does not occur on MacConkey agar, Marine agar, Tryptic Soy agar, or Luria Bertani agar. In the API 20NE strip, it was positive for esculin hydrolysis, gelatin hydrolysis, and β-galactosidase activities. In the API ZYM strip, alkaline phosphatase, leucine arylamidase, valine arylamidase, trypsin, α-Chymotrypsin, acid phosphate, naphthol phosphohydrolase, β-galactosidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, esterase, esterase lipase, cystine arylamidase, α-galactosidase, and α-flucosidase activities are present; lipase, β-glucuronidase, and α-mannosidase activities are absent. In the API 32GN kit, the following compounds are assimilated as sole carbon sources: glucose, melibiose, 2-ketogluconate, 3-hydroxybutyrate, L-alanine, sucrose, maltose, malonate, L-ribinose, inositol, L-sorbose, glycogen, and weakly assimilated propionate, acetate, lactate, valerate, L-serine, L-proline, N-acetylglucosamine, mannitol, and salicin; the other substrates such as caprate, citrate, 3-hydroxybenzonate, L-histidine, L-rhamnose, itaconate, suberate, L-arabinose, L-fucose, 5-ketogluconate, and 4-hydroxybenzoate are not assimilated. The major cellular fatty acids are C15:0 iso, C15:1 iso G and C17:0 iso 3OH. The only major respiratory quinone in strain BT541 is MK-7. The polar lipids are a phosphatidylethanolamine, three unidentified amino lipids, three unidentified aminophospholipids, an unidentified phospholipid, and five unidentified lipids. The strain BT-541 (KCTC 82123= JCM 37057) was isolated from a soil sample in South Korea.
The dimensions of the cells range from approximately 1-5 to 3 µm in length and 0.4-0.5 µm in diameter. After three days of incubation at 30 °C, colonies of strain BT-541 on R2A agar appear orange, smooth, convex, circular with smooth edges, and measure around 1 mm in diameter. Growth is observed at temperatures between 10-37 °C, with an optimum at 30 °C, and at pH levels of 6-10, with the ideal pH being 7.0, while it does not tolerate NaCl. This strain can grow on R2A agar, nutrient agar, and 1/10 tryptic soy agar. However, there is no growth on MacConkey agar, Marine agar, Tryptic Soy agar, or Luria Bertani agar. In the API 20NE strip tests, it showed positive results for esculin hydrolysis, gelatin hydrolysis, and β-galactosidase activity. In the API ZYM strip, activities for alkaline phosphatase, leucine arylamidase, valine arylamidase, trypsin, α-Chymotrypsin, acid phosphatase, naphthol phosphohydrolase, β-galactosidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, esterase, esterase lipase, cystine arylamidase, α-galactosidase, and α-flucosidase are present; however, lipase, β-glucuronidase, and α-mannosidase activities are not detected. In the API 32GN kit, the following substances can be utilized as the only carbon sources: glucose, melibiose, 2-ketogluconate, 3-hydroxybutyrate, L-alanine, sucrose, maltose, malonate, L-ribinose, inositol, L-sorbose, glycogen, along with weak assimilation of propionate, acetate, lactate, valerate, L-serine, and L-proline, N-acetylglucosamine, mannitol, and salicin are the only assimilated substrates; other substrates such as caprate, citrate, 3-hydroxybenzonate, L-histidine, L-rhamnose, itaconate, suberate, L-arabinose, L-fucose, 5-ketogluconate, and 4-hydroxybenzoate are not utilized. The major cellular fatty acids are C15:0 iso, C15:1 iso G, and C17:0 iso 3OH. The sole major respiratory quinone present in strain BT541 is MK-7. The polar lipids consist of a phosphatidylethanolamine, three unidentified amino lipids, three unidentified aminophospholipids, one unidentified phospholipid, and five unidentified lipids. The strain BT-541 (KCTC 82123= JCM 37057) was isolated from a soil sample collected in South Korea.
This work was supported by the Department of Biotechnology Research, Kyaukse, Ministry of Science and Technology, Myanmar, by the Department of Plant Medicine, Kyungpook National University, Daegu, South Korea, and by the International Scholar Exchange Fellowship program, Chey Institute of Advanced Studies.
None.
I declare that the manuscript is original, has not been published before and is not currently being considered for publication anywhere. We know no conflict of interest associated with this publication, and there has been no significant financial support for this work that could have influenced its outcomes.

©2025 San, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.