Journal of
eISSN: 2572-8466


Research Article Volume 11 Issue 5
1Department of Plant Pathology, Kerala Agricultural University, India
2Assistant Professor, Kerala Agricultural University, India
3Department of Vegetable Science, College of Agriculture, Kerala Agricultural University, India
Correspondence: M Vasantha, M.Sc student, Department of Plant Pathology, College of Agriculture, Vellanikkara, Kerala Agricultural University, Thrissur, Kerala, 680656, India
Received: September 21, 2024 | Published: October 14, 2024
Citation: Vasantha M, Mary CFG, Varghese SG, et al. Morpho-molecular characterization and in vitro management of bacterial wilt pathogen in cucurbits. J Appl Biotechnol Bioeng. 2024;11(5):159-168. DOI: 10.15406/jabb.2024.11.00373
Purposive sampling survey was conducted in 11 locations of three districts in Kerala and nine wilted cucurbits samples were collected. Isolation was carried on selective media Triphenyl Tetrazolium Chloride (TZC) agar medium and bacterial pathogen was identified based on cultural, morphological, biochemical characters and molecular characterization by 16S rRNA analysis. Studies showed that among nine cucurbits isolates obtained during survey, only three isolates proved the Koch’s postulates with per cent diseases incidence of 12.5 to 80 per cent. In three tested isolates a species-specific band of 1.5 kb was amplified in PCR that confirmed the identity of Klebsiella sp. However, this is the first report showing the Klebsiella sp. causing bacterial wilt on cucurbit crops in Kerala.
Keywords: bacterial wilt, characterization, molecular characterization, 16s rRNA, in vitro studies, ralstonia solanacearum, klebsiella sp
TZC, triphenyl tetrazolium chloride; ITS, internal transcribed spacer; PDI, per cent diseases incidence; CPG, casamino acid peptone glucose, IP, incubation period; PWI, per cent wilt incidence; PGPM, plant growth promoting microorganisms
Bacterial wilt disease causes significant yield loss in several crop plants. Among the different pathogens causing the bacterial wilt, Ralstonia solanacearum is the predominant one. The first record of bacterial wilt caused by R. solanacearum syn.1 Pseudomonas solanacearum (Smith) in the world was on potato by Burril in 1890 in Japan. R. solanacearum is considered one of the most destructive plant pathogens in tropical, subtropical and temperate regions worldwide, with the exception of the Antarctic continent.2 R. solanacearum1 causes bacterial wilt in more than 450 species of host plants belonging to approximately 54 botanical families.3
Mathews et al,.4 reported incidence of bacterial wilt in pumpkin and snake gourd from Kerala. The first record of occurrence of bacterial wilt of loofah [Luffa aegyptiaca] caused by R. solanacearum belonging to race I and biovar 3 was reported in India.5 Occurrence of bacterial wilt caused by R. solanacearum on bitter gourd and ridge gourd was first reported in India by Mathew et al,.4. Wicker et al,.6 reported the first record of bacterial wilt caused by R. solanacearum on cantaloupe [Cucumis melo], zucchini [Cucumis pepo], pumpkin [Cucumis sativus] and cucumber [Cucumis sativus] in Martinique (French West Indies). She et al,.7 reported bacterial wilt disease on Cucurbita maxima in Guangdong, China, is caused by R. solanacearum. Bitter gourd (Momordica charantia L.), was reported as a new host of R. solanacearum in Sri Lanka, causing loss in yield.8
In China, Ho et al,.9 reported the first incidence of Klebsiella pneumoniae sp. pneumoniae as the causal agent of bacterial leaf spot in banana. Recently, a causal agent of bacterial top rot in maize was reported as K. pneumoniae KpC4, which had been seen in Yunnan province of China.10 Klebsiella pneumoniae strain Borkar was reported as the causal agent of root bark necrosis and wilt in pomegranate plants.11
The symptoms caused by R. solanacearum can differ across wide range of susceptible hosts, but there are general symptoms shared among these hosts, such as the wilting and yellowing of younger leaves. Over time, the entire plant may rapidly wither and dry up, followed by yellowing of the foliage which ultimately leads to death of the plant. The vascular bundles in wilt-infected tomato plants exhibit long and narrow appearance dark brown streaks. Rawy et al isolated K. pneumoniae colonies using MacConkey agar. The colonies were distinguished by its mucoid growth and pink color.
In the past, bacterial pathogen was primarily identified and characterized based on their biochemical traits, but nowadays, genotypic methods have taken precedence. Among these methods, the comparison of the bacterial 16S rDNA gene sequence has become a preferred genetic technique. This preference arises from the fact that the 16S rDNA gene is a conserved region in the bacterial genome. Prior et al,.12 reported that, R. solanacearum strains are highly variable and adaptable with worldwide distribution and a large and expanding host range. They presented a classification system based upon phylogenetic analysis of sequence data generated from the 16S and 23S internal transcribed spacer (ITS) region, the endoglucanase gene and the mutS gene. She et al,.7 characterized 24 bacterial isolates from the diseased C. maxima using the universal PCR primers 27F and 1491R, each generated a 1.423 kb 16S rRNA gene fragment. BLASTn analysis of these 16S rRNA gene sequences revealed that they shared 100% identity with R. solanacearum strain GMI1000.
The identification of a novel Klebsiella species, designated as K. variicola, included total DNA-DNA hybridization, the observed monophyly in phylogenetic analysis derived from specific gene sequences (rpoB, gyrA, mdh, infB, phoE, and nifH), and distinctive phenotypic traits. Notably, the bacterial strains belonging to this newly proposed species exhibit genetic isolation from K. pneumoniae strains, lack the ability to ferment adonitol, and have been isolated from diverse plant sources such as banana, rice, sugar cane, and maize.13
Various management measures have been found effective against the pathogens R. solanacearum. Yuliar et al,.14 recommended the use of integrated biological control agents and organic matter for managing R. solanacearum. Two bacterial strains, Bacillus velezensis and Pseudomonas fluorescens were found efficient in controlling wilt caused by R. solanacearum in tomato under green house and field conditions.15 The bacterial wilt disease poses serious threat to cucurbit cultivation due to its fast spreading nature and difficulty in management. A thorough knowledge on etiology of the disease and pathogen characterization is necessary for formulating efficient management measures. With this view, the present research was undertaken to identify, characterize and develop suitable management measures against the bacterial pathogen.
Survey and sample collection
Purposive sampling survey was undertaken October, 2022 to August, 2023 to collect samples of cucurbit crops affected by bacterial wilt. The survey locations encompassed Thrissur, Malappuram and Palakkad districts, representing four distinct agro-ecological units (AEUs) in Kerala: North central laterites, Northern laterites, Palakkad central plains and Palakkad Eastern plains of central zone. The per cent disease incidence (PDI) of bacterial wilt in survey location was worked out by dividing the number of infected plants by the total number of plants and multiplied by 100.16
Symptomatology
Symptomatology studies were conducted under field conditions and typical wilt symptoms produced on cucurbit crops by the bacterial pathogen were observed and recorded in different survey locations of Kerala.
Isolation and maintenance of bacterial pathogen
The basal stem portion of wilted plants was washed under running tap water, followed by surface sterilization with 70 per cent alcohol. Afterward, the stems were cut into small pieces measuring 10-15 mm in length using a sterile blade. These pieces were then suspended in 5 ml of sterile water for 15-20 minutes. Once the water become cloudy with bacterial ooze, a small amount of the suspension was streaked onto Triphenyl Tetrazolium Chloride (TZC) agar as described by Kelman in 1954 with sterile inoculation loop. The plates were then incubated at room temperature for 48 hours. Single colonies with a round to oval shape and a red or pink centre were selected from the TZC agar plates. These selected colonies were purified by quadrant streaking on the same medium. Three to five characteristic single colonies were picked from purified plates and suspended in 5 ml of sterile distilled water in screw-capped plastic vials and stored in refrigerator at 4oC for future use. The virulence of the isolates was maintained by streaking on TZC agar once in every two months.
Cataloguing of bacterial pathogen
The bacterial pathogens isolated from the infected samples were systematically catalogued using abbreviations that denote information about the surveyed district, specific survey location and type of cucurbit affected. The districts of the purposive sampling survey were represented by the following abbreviations: T for Thrissur, P for Palakkad, and M for Malappuram. The specific survey locations within each district were designated as Vk (Vellanikkara), Ko (Kodakara), Pt (Pattambi), El (Elevanchery) and Pu (Puthur). The type of cucurbit crop affected by each isolate was denoted using the following abbreviations like B (Bitter gourd), P (Pumpkin), S (Snake gourd) and C (Salad cucumber). This systematic abbreviation system allowed for clear and concise identification of the bacterial isolates based on their origin and the type of cucurbit affected.
Pathogenicity studies
Pathogenicity of the isolates was conducted on respective cucurbit plants at the four to six leaf stages. The cucurbits plants raised in polybags were maintained in the net house of Department of Plant Pathology for conducting the pathogenicity studies. Various cucurbit hosts like bitter gourd (Preethi), pumpkin (Ambili), snake gourd (Dragon) and salad cucumber (KPCH-1), were inoculated through different viz., soil drenching with wounding and stem puncturing.17 At least 10 cucurbits seedling with three replication and control plants were maintained for the experiment.
Soil drenching with wounding
For conducting pathogenicity, the lateral roots of the young cucurbit seedlings were incised by inserting a knife at 1.5 cm away from the base of the plants, cutting to a depth of approximately 4 cm. Subsequently, 30 ml of a bacterial inoculum grown overnight in Casamino acid Peptone Glucose (CPG) broth with an optical density of 0.8-1 at OD600 and a concentration of 1 × 108 cfu/ml was poured onto the soil surrounding the base of the seedlings.
Stem puncturing
The stem puncturing was carried out by puncturing the stem of the cucurbit seedlings at the axis of the third fully expanded leaves from the apex with a needle dipped in inoculum and additionally a cotton swabbed with 100 μl of the bacterial suspension in CPG broth (OD600 = 0.8-1.0) was placed over the punctured site. The plants were maintained in the net house and were observed daily for up to 15 days for any wilt symptoms. The inoculated seedlings were maintained in the net house and observation and wilting was regularly recorded and per cent wilt incidence and incubation were worked out for each isolate separately on the hosts.
The incubation period (IP) and the per cent wilt incidence (PWI) were calculated by the formula to assess the virulence of the bacterial isolates.18
Incubation period (IP) = Number of days taken by the inoculated plants to produce visible symptoms.
Characterization of bacterial pathogen
The isolates of the bacterial wilt pathogen were characterized using a combination of cultural, morphological and biochemical characters. This multifaceted approach involved assessing various aspects of the bacterial pathogen characteristics to gain a comprehensive understanding of its identity and properties.
Cultural characterisation
The cultural characteristics of the isolates were studied by streaking the bacterium on TZC agar and MacConkey agar. A loopful of bacterial inoculum was streaked onto sterile molten TZC agar and MacConkey agar and incubated for 48 h. to allow the development of distinct, individual colonies. The colony traits, including size, pigmentation, shape, margin, fluidity and elevation, were carefully observed and documented according to the Bergey's Manual of systematic bacteriology.19
Morphological characterization
The morphological characterization of the bacterium was done by Gram staining and electron microscopy studies. These techniques provided detailed insights into the size, shape and structural features of pathogenic bacterium.
Gram staining
Gram staining is a widely used laboratory technique for categorizing bacteria based on their cell wall characteristics. Under sterile conditions, a drop of sterile distilled water was placed in the center of a clean glass slide. A loopful of bacterial inoculum was taken from a 48-hour-old bacterial culture and mixed thoroughly with two to three drops of sterile water, creating a thin bacterial smear on the glass slide. The smear was allowed to air dry and then heat fixed by passing the slide gently over a flame to fix the bacterial cells. The heat-fixed bacterial smear was flooded with Crystal Violet stain (the primary stain) for 1 minute, followed by gentle washing under running tap water. Next, the bacterial smear was flooded with iodine solution (mordant) for 1 minute, then washed with 95% alcohol (decolorizer) and rinsed under running tap water. Safranin (counterstain) was applied to the bacterial smear and left for 1 minute, followed by washing again under running tap water. Finally, the stained and air-dried sample on the glass slide was observed under oil immersion at 100 X magnification microscope.
Scanning electron microscopy
The detailed morphology of bacterial cell structure was studied by a scanning electron microscope (Tescan Vega-3 LMU) at the Central Instruments Laboratory of Kerala Veterinary and Animal Sciences University in Mannuthy, Kerala. The procedure followed was slight modification of the protocol described by Fischer et al,.20 Bacterial cells suspended in 1.5 ml of sterile water, harvested from a 24-hour-old culture, were centrifuged at 5000 rpm for 5 minutes. The supernatant was removed, and 1.5 ml of PBS (pH=7) was added. This mixture was centrifuged again at 5000 rpm for 5 minutes. The resulting pellet underwent serial dehydration with ethanol at concentrations of 30%, 50%, 70%, 80%, and 90%. After each ethanol dehydration step, the sample was incubated for 10 minutes and then centrifuged at 5000 rpm for 5 minutes. Subsequently, 1.5 ml of 100% ethanol was added to the pellet, which was then vortexed and incubated for 1 hour. After centrifugation at 5000 rpm for 5 minutes, the supernatant was discarded, and the pellets were dissolved in a small quantity of 100% ethanol. The sample was smeared on a stub and sputter coated with gold before being loaded into the scanning electron microscope.
The ideal working distance for the electron beam, which yields high-resolution images of bacteria, was determined through a trial-and-error approach. A high-resolution image of the bacterial cells were observed through a scanning electron microscope (SEM) with a working distance of 4.00 mm, 5.03 mm and 7.01 mm, resulting in a magnification of 15.0 kx and 25.1 kx.
Biochemical characterization
Accurate identification of bacteria requires polyphasic methodology based on morphological and biochemical data complemented with classical approach of 16S rRNA sequencing based molecular characterization.
Catalase test
A sterile glass slide was used, and a few drops of hydrogen peroxide (H2O2) were added to the center of the slide. Then, a loopful of bacterial inoculum was mixed into the solution. The formation of air bubbles in this mixture indicated a positive reaction.21
Oxidase test
An oxidase disc was placed upon a clean glass slide. A small amount of a 48-hour-old bacterial culture was then spreaded onto the disc using a sterile inoculation loop. Any color change on the oxidase disc was observed within 10 seconds.22
Starch hydrolysis/ Amylase production test
The sterilized starch agar medium was poured into sterile Petri plates and left to solidify. Bacterial isolates preserved under refrigerated condition were streaked onto the plates with starch agar medium and then incubated at 37°C for 48 hours. Following incubation, the plate's surface was flooded with iodine solution using a dropper for duration of 30 seconds, and any excess iodine was removed from the plate. The clear zone around the line of bacterial growth indicated a positive reaction.23
Gelatin hydrolysis
Bacterial isolates were stab-inoculated into nutrient gelatin. An uninoculated agar deep tube served as a control, and all the tubes were incubated at 37°C for duration of 4-7 days. After incubation period, all the tubes were refrigerated at 4°C for 15 minutes. A positive reaction was indicated by the liquefaction of the medium.24
Casein hydrolysis
Bacterial isolates were separately streak-inoculated onto the skim milk agar medium, and a control without bacterial streak was maintained and incubated at 37°C. A clear zone around the bacterial growth indicated a positive reaction.24
Urease test
Urea agar medium was first prepared and sterilized. Once the medium had cooled 1 g of glucose and 6 ml of phenol red (0.2%) were added, followed by a one-hour steaming process. An aqueous solution of urea (20%) was then incorporated into the medium, and the resulting mixture was sterilized through filtration. Slants were created using this medium, and bacterial cultures were inoculated onto them. The slants were subsequently incubated at 37°C for a period ranging from 24 to 48 hours. Examine for the change in the colour of medium for urease production.25
Hydrogen sulphide production test
Bacterial isolates were streaked onto the sterilized SIM agar media through stab inoculation, and the tubes were then incubated at 37°C for 48 h. Following incubation, the tubes were inspected for the presence of black colour along the line of the stab inoculation.24
Indole production test
Each bacterial isolate was inoculated into the freshly prepared 1% of tryptone broth. A control was maintained without any inoculation. Following inoculation, the media with the bacterial isolates were incubated for 48 hours at 35°C. After the incubation period, 1ml of Kovac's reagent was added to the inoculated tubes and gently shaken. The time taken for the development of purple color, if any, was recorded.24
Methyl red test
Each sterilized test tube containing 5 ml of the MR broth was inoculated with the test bacterium and then incubated for 48 hours at 35°C. An uninoculated control was also included. After the incubation period, five drops of methyl red pH indicator were added to each test tube, and any color change was observed. The development of a red color was recorded as a positive reaction, while no change in color in the broth was considered a negative reaction.24
Voges proskauer's test
Sterilized VP broth was dispensed into test tubes, and the test bacterium was inoculated into these tubes. They were then incubated for 48 hours at 35°C, alongside an uninoculated control. After the incubation, 12 drop of VP reagent I and two to three drops of VP reagent II were added to each tube. The tubes were gently shaken for 30 seconds without inserting cotton plugs to allow the bacterial cells to be exposed to oxygen. Following proper shaking, the test tubes were left undisturbed for 15 minutes to complete the reaction, and any color change was observed. The development of a pink or purple color was considered a positive reaction.24
Citrate fermentation test
Slants of Simmond's citrate agar were prepared and inoculated with bacterial isolates. An uninoculated slant was maintained as a control. The tubes were then left incubated at room temperature and monitored for any changes in color.26
Nitrate reduction test
Nitrate broth was prepared in test tubes and autoclaved. Inoculated bacterial culture into it and incubated for 18-24 hours. After incubation, added Nitrate reagents A (sulfanilic acid) and B (alpha-naphthylamine) sequentially, and observed for the development of a red color. A positive result indicated nitrate reduction, while a negative result suggests no nitrate reduction occurred.27
Lipid hydrolysis
To test for lipid hydrolysis, bacterial culture was streaked onto a tributyrin agar plate and incubated at the appropriate temperature (typically 35-37°C) for 24 to 48 hours. After incubation, examined the plate for a clear zone or halo around the bacterial growth, indicated the breakdown of tributyrin and a positive result for lipid hydrolysis.
Solubility in 3% KOH
A freshly prepared three percent potassium hydroxide solution was dispensed onto a clean glass slide using a dropper, typically in a quantity of three to four drops. A loopful of bacterial culture was collected using an inoculation loop and then mixed with the potassium hydroxide solution. After thorough mixing, the loop was gently lifted, and the slide was observed for the presence of fine threads of bacteria clinging to the loop.28
Molecular characterization
Bacterial isolates obtained from the diseased samples were further identified at the species level by 16S rRNA analysis. Bacterial genomic DNA was isolated utilizing the QIAGEN DNeasy UltraClean Microbial Kit (Cat. No. / ID: 12224-50), whereby a fragment of the 16S rRNA gene was subsequently amplified using specific primers, 16SrRNA-F and 16SrRNA-R. Following amplification, the resulting PCR amplicon underwent purification to eliminate contaminants, employing the QIAGEN QIAquick PCR Purification Kit (Cat. No. / ID: 28104). The quality of the isolated DNA was meticulously evaluated through agarose gel electrophoresis. For sequencing, the PCR products were subjected to the Big Dye™ Terminator v3.1 Cycle sequencing kit within an Applied Biosystems™ MiniAmp™ Plus Thermal cycler, following a designated PCR amplification profile consisting of an initial denaturation at 95°C for 3 minutes, followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 50°C for 30 seconds, and extension at 72°C for 45 seconds, with a final extension at 72°C for 3 minutes. Subsequently, the amplified products were analyzed on an ABI 3730xl Genetic Analyzer. Furthermore, an in silico analysis of the sequences was performed using the nucleotide Basic Local Alignment Search Tool (BLASTn) software, facilitating a thorough comparison with sequences cataloged in the NCBI database to ascertain their homology.
In vitro evaluation of antibacterial antibiotics and biocontrol agents
In vitro evaluation of antibacterial antibiotics against isolated bacterial pathogen
Antibacterial antibiotics such as Streptocycline and 2-Bromo-2-nitropropane-1,3-diol (Bronopol) were assessed at @ 200, 250, 300 ppm concentrations to determine their effectiveness in inhibiting the proliferation of isolated bacterial pathogens. The evaluation was carried out using the inhibition zone assay method outlined by Bauer et al. A loopful of a 48-hour-old bacterial culture was inoculated into 20 ml of CPG broth in 100 ml conical flask and placed in a shaker at 37°C, with agitation at 120 rpm, for three days. After this incubation period, 10 ml of a dense bacterial suspension was aseptically transferred to 100 ml of CPG agar medium, which had been melted and cooled, with constant stirring. This thoroughly mixed medium was then poured into sterile Petri plates and allowed to solidify. To assess the effectiveness of different concentrations of antibacterial antibiotics, a specific amount of antibiotic was mixed with 100 ml of sterile distilled water. Wells were created in the agar using 8 mm cork borer, and 50 µl of each antibiotic solution was added to separate wells, while a well loaded with sterile distilled water served as a control. This experiment was performed in triplicate under sterile conditions. Subsequently, the plates were incubated at 28°C and examined 24 hours post-incubation for the presence of clear zones around the wells. The diameter of these inhibition zones was measured in millimeters.
In vitro evaluation of biocontrol agents against bacterial pathogen
Biocontrol agents, including Trichoderma asperellum (KAU reference culture), Pseudomonas fluorescens (KAU reference culture) and Plant Growth Promoting Microorganisms (PGPM) at a concentration of 2%, were evaluated for their effectiveness in suppressing the growth of isolated bacterial pathogens.
In-vitro evaluation of Trichoderma asperellum against bacterial pathogen
The antagonistic potential of Trichoderma asperellum against isolated bacterial pathogens was assessed using the dual culture method. The isolated bacteria were streaked at 2 cm from the periphery Petri plates containing PDA agar. A mycelial disc of 8 mm in diameter was taken from an actively growing culture of species was positioned on the opposite side 2 cm from the periphery of the plates. These plates were then incubated at 28 ± 2°C for 2-3 days. The extent of antagonistic activity by the biocontrol agent was determined by measuring the growth of the bacterial pathogen in the dual culture plate and in control plate. The per cent inhibition (PI) of pathogen was calculated using the formula (Vincent 1974).
In vitro evaluation of Pseudomonas fluorescens against bacterial pathogen
In vitro study aimed at assessing the effectiveness of antagonistic Pseudomonas fluorescens was conducted using the cross-streaking method. The antagonist was inoculated by a single stripe in the middle of nutrient agar medium and after an interval of time; the plate was inoculated with bacterial isolate by triple stripe, perpendicular to the central stripe.29 While the medium streaked only with bacterial isolate serves as control. All the plates were subsequently incubated at 37°C for 72 hours, and the measurement of the inhibition zone created around the bacterial pathogen by the antagonistic Pseudomonas fluorescens was recorded.
In vitro evaluation of PGPM against bacterial pathogen
The PGPM formulation was assessed for their antagonistic effect against isolated bacterial pathogen using poison food method. Initially, two grams of PGPM formulation were aseptically combined with 100 ml of precooled PDA medium taken in a 250 ml conical flask, as per the recommended dosage of 2%, and mixed thoroughly to prepare uniform suspension and pouring into sterile Petri plates for solidification. The bacterial pathogen was spot inoculated on the poisoned medium and control plates without PGPM were also maintained. Both the treatment and control plates were incubated at room temperature for 48 hours, and observations concerning the development of inhibition zones were recorded and analyzed.
Isolation and maintenance of bacterial pathogen
Cucurbit plants which exhibited typical bacterial wilt symptoms were collected during purposive sampling survey and brought to laboratory for isolation of pathogen. To confirm the presence of bacteria, wilted plants were subjected to ooze test. The ooze test yielded fine milky white ooze with mass of bacteria streaming out from the margins of the cut end, thus confirming the presence of bacteria in the plant sample.
The bacterial wilt pathogen was isolated from infected cucurbits plants on Triphenyl Tetrazolium Chloride (TZC) agar medium (Kelman 1954). Creamy white, irregular, smooth, convex, fluidal and slimy colonies with pink to reddish centre were observed in isolation plate after 24-48 h of incubation (Figure 1). The pathogen was purified by repeated quadrant streaking on TZC medium. The purified bacterial culture was preserved in sterile water in screw capped vials and stored under refrigerated conditions at 4oC for further studies. The virulence of the isolates was maintained by streaking on TZC agar once in two months.
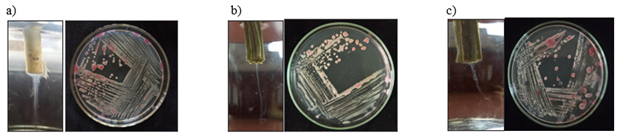
Figure 1 Isolation of bacterial pathogen on TZC agar medium. a) TVkP-1 isolate; b) PPtP isolate; c) PElB isolate.
Pathogenicity studies
Among the nine isolates tested for pathogenicity (TKoB, TVkP-1, PPtP, TVkC, PElB, PElS, TVkB, TPuB, and TVkP-2) on their respective cucurbit host and specific variety, only three isolates viz., TVkP-1, PPtP on pumpkin and PElB on bitter gourd established the pathogenicity with 12.5 per cent, 16.66 per cent and 80 per cent wilt incidence respectively (Table 1). Typical wilt symptoms were observed on healthy plants inoculated with different isolates of bacterial pathogen. The preliminary diagnosis of wilted plants was carried out by ooze test and the pathogen was re-isolated from the wilted plants. The colonies produced on TZC agar were compared with the original colonies and found similar. Among the nine isolates only three isolates, TVkP-1, PPtP and PElB which confirmed pathogenicity were selected for further characterization of pathogen.
|
Sl. No. |
Type of cucurbit crop |
Isolate |
Incubation period (days) |
Reisolation (+/-) |
Percent disease incidence (%) |
|
1 |
Bitter gourd |
TKoB |
- |
- |
- |
|
2 |
Pumpkin |
TVkP-1 |
10 |
+ |
12.5 |
|
3 |
Pumpkin |
PPtP |
10 |
+ |
16.66 |
|
4 |
Bitter gourd |
PElB |
6 |
+ |
80 |
|
5 |
Snake gourd |
PElS |
- |
- |
- |
|
6 |
Salad cucumber |
TVkC |
- |
- |
- |
|
7 |
Bitter gourd |
TVkB |
- |
- |
- |
|
8 |
Pumpkin |
TVkP-2 |
- |
- |
- |
|
9 |
Bitter gourd |
TPuB |
- |
- |
- |
Table 1 Details of pathogenicity studies by different isolates on cucurbit crops
Cultural and morphological characterization of the bacterial pathogen
The bacterial colonies of three isolates, TVkP-1, PPtP and PElB were irregular, small to moderately sized and appeared creamy white with light pink pigment in the centre (Table 2). Produced highly fluidal colonies with entire margin and convex elevation. The bacterial characters recorded on MacConkey were small, circular, pink-coloured mucoid colonies. All the three bacterial isolates, (TVkP-1, PPtP and PElB) appeared pink on Gram staining which confirmed that all isolates were Gram negative (Figure 2). The SEM magnification and working distance of the electron beam were adjusted by trial and error method so as to acquire high resolution images, 5.03 mm for TVkP-1 and 7.01 mm for PPtP and PElB isolates (Figure 3). Typical rod-shaped cells of all the bacterial isolates with size ranging from 1.47 μm x 0.55 μm to 1.89 μm x 0.6μm were observed.
|
Sl. No. |
Isolate |
Size |
Pigmentation |
Form |
Margin |
Elevation |
Fluidity |
|
1 |
TVkP-1 |
Small to moderate |
Creamy white with pinkish centre |
Irregular |
Entire |
Convex |
Low |
|
2 |
PPtP |
Moderate to large |
Creamy white with dark pinkish centre |
Irregular |
Entire |
Convex |
Moderate |
|
3 |
PElB |
Small to moderate |
Creamy white with light pink centre |
Irregular |
Entire |
Convex |
High |
Table 2 Cultural characteristics of the bacterial isolates on TZC agar
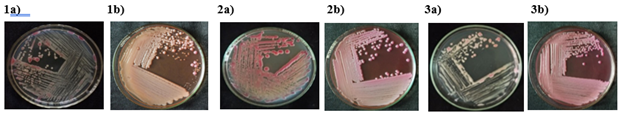
Figure 2 Cultural characterization of bacterial pathogen on TZC agar and MacConkey agar. 1) TVkP-1 isolate colony characters on a) TZC agar and on b) MacConkey agar,2) PPtP isolate colony morphology on a) TZC agarand on b) MacConkey agar, 3) PElB isolate colony morphology on a) TZC agarand on b) MacConkey agar.
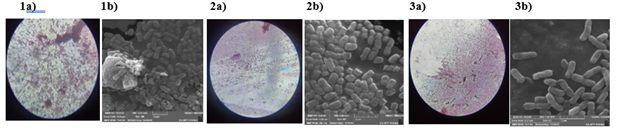
Figure 3 Morphological characterization of bacterial pathogen by Gram staining and Scanning electron microscopy. 1a) and 1b) TVkP-1 isolate; 2a) and 2b) PPtP isolate; 3a) and 3b) PElB isolate.
Biochemical characterization of the bacterial pathogen
All the three isolates associated with cucurbits bacterial wilt showed positive results for starch hydrolysis, urease test, MR test, citrate utilization, catalase test, oxidase test, nitrate test and motility test whereas the bacterial isolates were negative to gelatin liquefaction, casein hydrolysis, H2S test, indole production, VP test and lipid test (Figure 4). Based on the biochemical test results, the bacterial isolates showed similarity to Ralstonia solanaccearum of Burkholderiaceae as well to Klebsiella sp. of family Enterobacteriacea. The positive results of the motility test deviated from that of Klebsiella sp. since the genera is non motile and shows negative reaction (Table 3). Hence further characterization based on the amplification of the 16S rRNA of the bacterial pathogen was carried out for confirming the identity of the pathogen.
|
Sl. No. |
Biochemical test |
Reaction of isolates |
||
|
TVkP-1 |
PPtP |
PElB |
||
|
1 |
Starch hydrolysis |
+ |
+ |
+ |
|
2 |
Gelatin liquefaction |
- |
- |
- |
|
3 |
Casein hydrolysis |
- |
- |
- |
|
4 |
Urease test |
+ |
+ |
+ |
|
5 |
H2S test |
- |
- |
- |
|
6 |
Indole production |
- |
- |
- |
|
7 |
MR test |
+ |
+ |
+ |
|
8 |
VP test |
- |
- |
- |
|
9 |
Citrate utilization |
+ |
+ |
+ |
|
10 |
Catalase test |
+ |
+ |
+ |
|
11 |
KOH test |
+ |
+ |
+ |
|
12 |
Oxidase test |
+ |
+ |
+ |
|
13 |
Nitrite test |
+ |
+ |
+ |
|
14 |
Lipase test |
- |
- |
- |
|
15 |
Motility test |
+ |
+ |
+ |
Table 3 Biochemical characteristics of bacterial isolates
+ positive; - negative
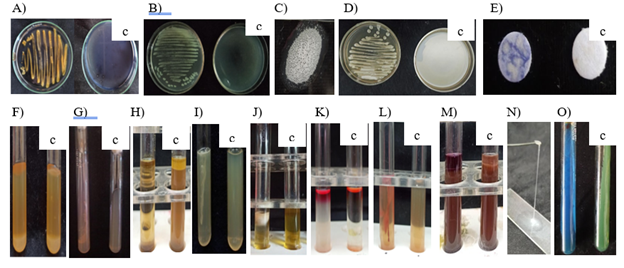
Figure 4 Biochemical characterization of bacterial pathogen: A: Starch hydrolysis test (+), B: Lipid hydrolysis test (-), C: Casein hydrolysis test (-), D: Oxidase test (+), E: Catalase test (+), F: Gelatin liquefaction(-), G: Urease test (+), H: Indole production test (-), I: H2S production test (-), J: VP test (-), K: MR test (+), L: Motility test (+), M: Nitrate reduction test (+), N: KOH test (+), O: Citrate test (+), c: Control
Molecular characterization of bacteria was conducted by amplification and sequencing of 16S rRNA region of bacterial genomic DNA. The PCR amplification of all the three bacterial isolates, TVkP-1, PPtP and PElB using bacterial universal primers which produced approximately 1.5 kb clear band (Figure 5). The isolate TVkP-1 causing wilt in pumpkin was characterized by amplification and sequencing of 16S rRNA region using two different sets of primers viz., 1550 F and 1550 R, and 16SrRNA-F-27F and 16SrRNA-R-1492R primers. The results obtained by amplifying and sequencing 16SrRNA region of bacterial genome using 1550 F and 1550 R primers yielded 97.53 per cent sequence similarity with Klebsiella quasipneumoniae subsp. quasipneumoniae strain L4 with 92 per cent query cover and submitted the sequences in NCBI, GenBank (accession no.: PP439468). And other primers set, 16SrRNA-F-27F and 16SrRNA-R-1492R were also employed for the amplification and subsequent sequencing of the 16S rRNA region of bacterial genomic DNA. The BLASTn analysis revealed that TVkP-1 exhibited 92.26 per cent identity with Klebsiella pneumoniae strain Kpn21661 with query coverage of 84 per cent. The isolate PPtP from pumpkin, was characterized by amplification and sequencing of the 16S rRNA gene using primers 16SrRNA-F-27F and 16SrRNA-R-1492R. BLASTn analysis revealed 99.55 per cent identity with Klebsiella pneumoniae strain zg2010 with 98 per cent query coverage and submitted the sequences in NCBI, GenBank (accession no.: PP439984). The isolate PElB associated with wilting in bitter gourd was identified by amplification and sequencing of 16S rRNA gene using primers 27F and 1492R. BLASTn analysis showed 100 per cent identity with Klebsiella pneumoniae strain 2020C07 with query coverage of 99 per cent and submitted the sequences in NCBI, GenBank (accession no.: PP439836) (Table 4).
|
Bacterial isolate |
Pathogen identified |
Primers name |
Percent identity |
|
TVkP-1 |
quasipneumoniae subsp. quasipneumoniae strain L4 |
1550 F, 1550 R |
97.53 |
|
Klebsiella pneumoniae strain Kpn2166 |
16SrRNA-F-27F and |
92.26 |
|
|
PPtP |
Klebsiella pneumoniae strain zg2010 |
16SrRNA-R-1492R |
99.55 |
|
PElB |
Klebsiella pneumoniae strain 2020C07 |
|
100 |
Table 4 Molecular characterization of bacterial isolates using 16SrRNA with per cent identity
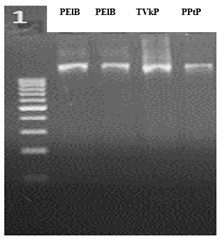
Figure 5 Gel band image of three bacterial isolates with 16S rRNA. 1st and 2nd lane PElB isolate, 3rd lane TVkP-1 isolate and 4th lane PPtP isolate.
In vitro evaluation of antibacterial antibiotics and biocontrol agents against the bacterial pathogen
In vitro evaluation of antibacterial antibiotics against bacterial pathogen
The two antibacterial antibiotics viz., Streptocycline and 2-Bromo-2-nitropropane-1,3-idol/ Bronopol at 200 ppm, 250 ppm and 300 ppm were tested under in vitro conditions for their efficiency in inhibiting the growth of bacterial pathogen. There was significant difference in the inhibition of bacterial growth by two antibacterial antibiotics tested against TVkP-1 under in vitro condition which ranged from 13.52 to 25.92 per cent with diameter of inhibition zones ranging from 12.2 to 23.3 mm (Table 5). Streptocycline showed maximum inhibition of 25.92 per cent with 23.3 mm diameter of inhibition zone at 300 ppm and was on par with inhibition at 250 ppm (24.81 %). Least inhibition of 16.47 per cent with 14.9 mm inhibition diameter was recorded at 200 ppm. The antibiotic Bronopol recorded maximum inhibition of 14.99 per cent with inhibition zone of 13.5 mm diameter at 300 ppm which was on par with 250 ppm (14.25%) and minimum inhibition of 13.52 per cent and 12.2 mm inhibition zone was observed at 200 ppm. In vitro efficacy of two antibacterial antibiotics tested at different concentrations against PPtP showed significant difference in their efficiency. Streptocycline at 300 ppm showed maximum inhibition of 25.55 per cent with 23 mm diameter of inhibition zone and least inhibition of 18.51 per cent with 16.7 mm diameter of inhibition at 200 ppm (Figure 6). The antibiotic Bronopol showed maximum per cent inhibition of 14.44 percent and 13 mm inhibition diameter at 300 ppm and minimum inhibition was observed at 250 ppm with 13.14 per cent with inhibition diameter of 11.8 mm. In vitro evaluation of antibacterial antibiotics against PElB isolates showed the inhibition ranging from 13.7 to 29.77 per cent and diameter of inhibition zones ranged between 12.3 to 26.8 mm (Figure 6).
|
Sl No. |
Antibiotics |
Conc. (ppm) |
TVkP-1 |
PPtP |
PElB |
|||
|
Inhibition zone(mm) |
*Percent inhibition |
Inhibition zone(mm) |
*Percent inhibition |
Inhibition zone(mm) |
*Percent inhibition |
|||
|
1 |
Streptocycline |
200 |
14.9 |
16.47 (4.12)b |
16.7 |
18.51 (4.36)c |
15.7 |
17.38 (4.23)c |
|
250 |
22.3 |
24.81 (5.03)a |
20.5 |
22.77 (4.82)b |
18.5 |
20.55 (4.59)b |
||
|
300 |
23.3 |
25.92 (5.14)a |
23 |
25.55 (5.10)a |
26.8 |
29.77 (5.5)a |
||
|
200 |
12.2 |
13.52 (3.74)d |
12.3 |
13.7 (3.77)e |
13.2 |
14.62 (3.89)de |
||
|
2 |
Bronopol |
250 |
12.8 |
14.25 (3.84)cd |
11.8 |
13.14 (3.69)e |
12.3 |
13.70 (3.77)e |
|
300 |
13.5 |
14.99 (3.94)c |
13 |
14.44 (3.87)d |
13.4 |
14.88 (3.93)d |
||
|
CV |
1.89 |
1.39 |
2.265 |
|||||
|
|
CD (0.05) |
|
|
0.125 |
|
0.092 |
|
0.151 |
Table 5 In vitro evaluation of antibacterial antibiotics against bacterial pathogen
*Mean value of three replications
In each column figure followed by the same letter do not differ significantly according to DMRT.
√X+ 0.5 transformed values are given in parentheses
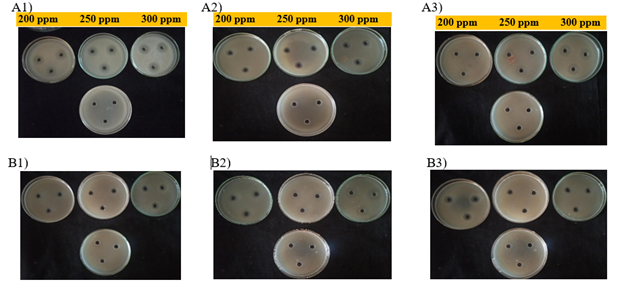
Figure 6 In vitro evaluation of antibacterial antibiotics against bacterial pathogen, Streptocycline (A1-A3) and Bronopol (B1-B3) 1) TVkP-1 isolate; 2) PPtP isolate; 3) PElB isolate.
In vitro evaluation of biocontrol agents against bacterial pathogen
The fungal antagonist Trichoderma asperellum (KAU reference culture) was evaluated against the selected bacterial isolates by dual culture assay. From the results it was evident that all the tested bacterial isolates showed different degree of inhibition ranging between 64.93 to 71.58 per cent, with a maximum (71.58 %) against TVkP-1 isolate followed by 68.31 per cent inhibition by PPtP. The lowest inhibition (64.93 %) was recorded for PElB (Figure 7). The bacterial antagonist Pseudomonas fluorescens (KAU reference culture) was evaluated against the bacterial isolates by inhibition zone assay. The in vitro studies conducted using Pseudomonas fluorescens against three bacterial isolates revealed that, the bacterial antagonist completely failed to inhibit the growth of all bacterial isolates without any lysis at the point of contact of cross streaking (Figure 7). In vitro evaluation of PGPM (Plant Growth Promoting Microorganisms) against the bacterial isolates was carried out by poison food technique (Figure 7). It was observed that PGPM inhibited the growth of bacterial isolates, TVkP-1 and PElB whereas no effect was observed against the bacterial isolate, PPtP (Table 6).
|
Sl. No. |
Antibiotics |
*Percent inhibition |
||
|
TVkP-1 |
PPtP |
PElB |
||
|
1 |
Trichoderma asperellum |
71.58 (8.49)a |
68.31 (8.30)a |
64.93 (8.089)a |
|
2 |
Pseudomonas fluorescens |
0 (1.81)b |
0(1.18)b |
0(1.81)b |
|
CV |
5.37 |
2.77 |
4.98 |
|
|
|
CD (0.05) |
0.32 |
0.17 |
0.29 |
Table 6 In vitro evaluation of biocontrol agents against bacterial pathogen
*Mean value of three replications
In each column figure followed by the same letter do not differ significantly according to DMRT.
√X+ 0.5 transformed values are given in parentheses
Managing bacterial wilt is challenging, due to fast spreading nature of bacteria. Bacterial wilt remains a persistent threat, demanding ongoing research for sustainable control in cucurbit cultivation. All the three isolates associated with cucurbits bacterial wilt showed positive results for starch hydrolysis, urease test, MR test, citrate utilization, catalase test, oxidase test, nitrate test and motility test whereas the bacteria was negative to gelatin liquefaction, casein hydrolysis, H2S test, indole production, VP test and lipid test. The biochemical tests are in conformity with Ralstonia solanacearum as reported by several workers Wicker et al,.6 Mathew et al.,4 and Rajapaksha et al,.8 The biochemical test results were also found similarity with reports of Klebsiella pneumoniae strain Borkar infecting pomegranate documented by Ajayasree et al.,11 and Klebsiella pneumoniae (KpC4) infecting maize by Huang et al,.30 It was found positive to urease test, starch hydrolysis, methyl red test, nitrate reduction, catalase test and citrate utilization, while it was negative for gelatin hydrolysis, indole and H2S production. Molecular characterization of bacteria was done for identification of pathogen of cucurbit wilt by amplification and sequencing of 16S ribosomal RNA region of bacterial genomic DNA. The pumpkin isolate, PPtP which was characterization through amplification and sequencing of the 16S rRNA gene, employing the primer set 16SrRNA-F-27F and 16SrRNA-R-1492R. BLASTn analysis displayed a high sequence identity of 99.55% with Klebsiella pneumoniae strain zg2010, identified by accession number JX435602.1, and query coverage of 98%. Ajayasree et al.,11 reported Klebsiella pneumoniae as a plant pathogenic bacterium causing disease in plants through the analysis of the 16S rRNA gene sequence, which had been deposited in the NCBI database under the Gene accession number KY941097.1. The wilt associated isolate, PElB in bitter gourd was recognised through the amplification and sequencing of the 16S rRNA gene, employing the primer set 27F and 1492R. NCBI database analysis indicated a complete sequence identity of 100% and query coverage of 99% with Klebsiella pneumoniae strain 2020C07. The corresponding accession number for this identical sequence was CP129799.1. Huang et al,.31 reported Klebsiella pneumoniae (KpC4) as top rot pathogen in maize using 16S rRNA primers which produced 1.5 kb amplicon and showed highest sequence homology with Klebsiella pneumoniae. Through the analysis of members of each Klebsiella sp. and several other Enterobacteriaceae members to study the 16S rDNA, revealed that when 16S rDNA similarity values exceed 97.0 ± 97.5%, relying solely on 16S rDNA sequencing may not be adequate for determining species identities or relationships. However, based on reviews of literature this is probably the first report of Klebsiella sp. on cucurbit crops. The antibacterial antibiotics tested against TVkP-1 under in vitro condition showed per cent inhibition ranging from 13.52 to 25.92 per cent with diameter of inhibition zones ranging from 12.2 to 23.3 mm. Streptocycline showed maximum inhibition of 25.92 per cent with 23.3 mm diameter of inhibition zone at 300 ppm and was similar to the inhibition at 250 ppm (24.81 %). The results are in agreement with the reports of Singh and Jagtap (2017) and Babu et al,.32 who found that Ralstonia solanacearum was sensitive to Streptocycline at higher concentration producing maximum inhibition zone. Least inhibition of 16.47 per cent with 14.9 mm inhibition diameter was recorded at 200 ppm. The antibiotic Bronopol recorded maximum inhibition of 14.99 per cent with inhibition zone of 13.5 mm diameter at 300 ppm which was on par with 250 ppm (14.25%) and minimum inhibition of 13.52 per cent and 12.2 mm inhibition zone was observed at 200 ppm. Similar results for Bronopol producing lowest inhibition zone was recorded by Gopalakrishnan et al. (2014) against R. solanacearum.
The results of in vitro efficacy of two antibacterial antibiotics tested at different concentrations against PPtP showed significant difference in their efficiency. Streptocycline at 300 ppm showed maximum inhibition of 25.55 per cent with 23 mm diameter of inhibition zone and least inhibition of 18.51 per cent with 16.7 mm diameter of inhibition at 200 ppm. The antibiotic Bronopol showed maximum per cent inhibition of 14.44 per cent and 13 mm inhibition diameter at 300 ppm and minimum inhibition was observed at 250 ppm with 13.14 per cent with inhibition diameter of 11.8 mm.
Similarly, in vitro evaluation of antibacterial antibiotics against PElB isolates showed the inhibition ranging from 13.7 to 29.77 per cent and diameter of inhibition zones ranged between 12.3 to 26.8 mm. Streptocycline showed maximum inhibition of 29.77 per cent with 26.8 mm inhibition zone at 300 ppm and least inhibition of 17.38 per cent with 15.7 mm inhibition diameter at 200 ppm. The antibiotic Bronopol showed highest inhibition of 14.99 per cent with 13.5 mm inhibition diameter at 300 ppm and least inhibition with diameter of inhibition of 13.70 per cent with 12.3 mm at 250 ppm. The research findings of Das et al,.33 revealed that Streptocycline showed an inhibition zone of 32.5 mm against K. pneumoniae. The bacterial antagonist T. asperellum was evaluated against the selected bacterial isolates by dual culture technique. The results indicated varying degrees of inhibition, ranging from 64.93% to 71.58%. The highest inhibition (71.58%) was observed against the TVkP-1 isolate, followed by 68.31% inhibition against PPtP. The lowest inhibition (64.93%) was recorded for PElB. The reduction in bacterial growth was evident in treatment plates after three days of incubation at the point where the pathogen and antagonist interact. After four days, bacterial growth completely ceased due to the overgrowth of the antagonist on the bacterial isolates. Within a week of incubation, the fungal antagonist had completely overgrown and lysed all bacterial isolates. Microscopic observations revealed thickened and swollen antagonist mycelium, along with disintegrated bacterial cells. Additionally, the colour of the bacteria changed from creamy white to yellow in all treatment plates. Konappa et al,.34 and Mohamed et al,.35 reported that the T. asperellum was efficient in inhibiting against R. solanacearum and enhanced yield of tomato and potato crops respectively. The bacterial antagonist, Pseudomonas fluorescens (KAU reference culture) was evaluated against selected bacterial isolates, TVkP-1, PPtP and PElB and found that the bacterial antagonist completely failed to inhibit the growth of the bacterial pathogen without any lysis at the point of contact of cross streaking.
This is the first report showing the Klebsiella sp. causing bacterial wilt on cucurbit crops in Kerala. Further characterization of bacterial wilt pathogen is needed to understand the etiology, mode of spread, and survival of pathogen, and identification of pathogen using species specific primers, and various antibacterial antibiotics should be tested against bacterial pathogen for efficient management of disease in field condition.36–38
In this article we have characterized the bacterial wilt pathogen causing disease on cucurbit crops in Kerala. Thus, based on pathogenicity, cultural, morphological, biochemical and molecular tests, this pathogen was identified as Klebsiella pneumonia sp. Ajayasree and Borkar (2018) reported that Klebsiella pneumoniae strain Borkar as the causal agent of root bark necrosis and wilt in pomegranate plants. However, this is the first report on this bacterial wilt pathogen of cucurbits from India. This study contains valuable information to identify Klebsiella species- specific or -related pathogenesis and enhance our present knowledge of the cucurbit bacterial wilt disease.
We thank Department of Plant Pathology, College of Agriculture, Vellanikkara, Kerala Agricultural University, Thrissur at Kerala for providing all the necessary resources for research work completion.
The authors declare no conflicts of interest.
This work was supported by Kerala Agricultural University under M.Sc. Research project Order No. R7/66317/2022.

©2024 Vasantha, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.