Journal of
eISSN: 2572-8466


Research Article Volume 3 Issue 5
Department of Bioinformatics, Central Agricultural University, India
Correspondence: Ajit Kumar Roy, Bioinformatics Center, College of Fisheries, Lembucherra, Tripura, Central Agricultural University, India
Received: June 12, 2017 | Published: August 29, 2017
Citation: Acharjee S, Upadhyay AD, Roy AK, et al. Insilico analysis of myostatin protein of Labeo calbasu . J Appl Biotechnol Bioeng. 2017;3(5):422-428. DOI: 10.15406/jabb.2017.03.00080
Myostatin is a protein released in autocrine fashion which exerts negative effect for skeletal muscle growth during myoblast proliferation and differentiation in teleost fishes. Myostatin is a member of the TGF-b family encoded by MSTN gene, also known as growth differentiation factor 8(GDF-8). MSTN gene knock-out exhibited hypertrophy and hyperplasia of skeletal muscle, but over expression or systemic administration of myostatin causes muscle atrophy. Blocking of myostatin activity helps in generating higher muscle growth rate and mutation in MSTN gene creates double-muscle phenotype. Therefore, insilico analysis of Myostatin in an Indian major carp species Labeo calbasu has carried out in the present study for understanding structural organization of the concerned protein. Amino acid sequence heaving the accession number AEN75195.1 was retrieved from NCBI. The sequence was subjected to Expasy ProtParam, PSIPRED and GORIV, PDB sum and SWISSMODEL/Workspace for finding out the physicochemical properties, secondary structure prediction and template selection procedure respectively. The predicted 3D structure of Myostatin by Swiss model server was validated through RAMPAGE server and then submitted to Protein Model Database (PMDB) heaving the ID No: PM0081037. The extracellular location was also analysed by Hum-mPLoc 2.0. The structural examination of Myostatin will help in development of antagonistic protein or inhibitor, which can shed a new direction for the growth promotion of piscine species during various periods of life cycle.
Keywords: myostatin, in silico, homology modeling, labeo calbasu, GORIV. expasy protparam, MSTN, uniprotKB
GDF, growth differentiation factor; PMDB, protein model database; NCBI, national center for biotechnology information; GRAVY, grand average of hydropathicity
Myostatin is a protein released from myocytes acting in autocrine fashion for inhibiting the myogenesis. It is a member of the TGF-b protein family encoded by MSTN gene which acts as a negative regulator for skeletal muscle growth during myoblast proliferation and differentiation in teleost fishes.1 Due to its involvement in growth processes it is also known as growth differentiation factor 8 (GDF-8). MSTN gene Knock out showed both the hypertrophy and hyperplasia of skeletal muscle,2 whereas, over expression or systemic administration of it resulted muscle atrophy.4 In agricultural and veterinary fields, treatment for blocking of myostatin activity is in practice which helps in generating higher muscle growth rate. Generally, it is expressed in skeletal muscle and the mutation in MSTN gene creates double-muscle phenotype2,5,6 in several domesticated species like Belgian blue and Piedmontese cattle7 or Texel sheep.8 However from fishes such as rainbow trout,9,10 atlantic salmon (Salmo salar),11 zebra fish (Danio rerio),12,13 sea bream (Sparus aurata),14 Orange spotted grouper (Epinephelus coioides),15 medaka (Oryzias latipes),16 baramundi (Lates calcarifes),17 sea perch (Lateolabrax japonicus)18 and catfish (Ictaluruspunctatus)19 etc the presence of Myostatin genes were well reported. Studies on myotomal musculature of fishes are interesting area of research since few decades. In vivo blocking of Myostatin gene expression in skeletal muscle of fishes was also carried out by Terova et al.20 The structural exploration of Myostatin in an Indian major carp species like Labeo calbasu has a potential significance, which can ultimately results to a conception of development of antagonistic protein or inhibitor protein for generating higher growth in fishes during different stages of life cycle.
Sequence retrieval
The amino acid sequence of Myostatin of Labeo calbasu was retrieved from National Center for Biotechnology Information (NCBI) heaving the accession number AEN75195.1. The FASTA format of the sequences were downloaded and used for further analysis. The retrieved sequence was verified by peptide search in the UniProtKB heaving the entry no. G3EIK5.
Physicochemical properties of myostatin
Expasy ProtParam server was used for determination of physicochemical properties of the Myostatin protein.21 The molecular weight, theoretical pI, the % of amino acid composition, total number of negative and positive residue, atomic composition, extinction coefficient, estimated half-life, instability index, aliphatic index, grand average of hydropathicity (GRAVY) were analyzed by this web tool.
Secondary structure prediction
Secondary structure of the Myostatin was analysed by PSIPRED22 and GORIV23 methodology.
Template selection
Template search for the query protein, Myostatin was performed through PDBsum database which presented 114 hits.24 The template 3hh2 (A) was selected showing 89% sequence identity. The template sequence was also cross verified by SWISSMODEL/Workspace which displayed 88.99% sequence identity with the query sequence.25
Homology modeling
By homology modeling, three dimensional structure of Myostatin was predicted using Swiss model server.26,27
Validation of the prediction
The predicted structure of the Myostatin was validated through Ramachandran plot by utilizing RAMPAGE server.28
Protein model database
The predicted model was also submitted to the Protein Model Database (PMDB). PMDB is a three dimensional protein models database.29
Subcellular localization
Hum-mPLoc 2.0 was used to predict the subcellular localization of the concerned protein Myostatin.30
Analysis of retrieved sequences
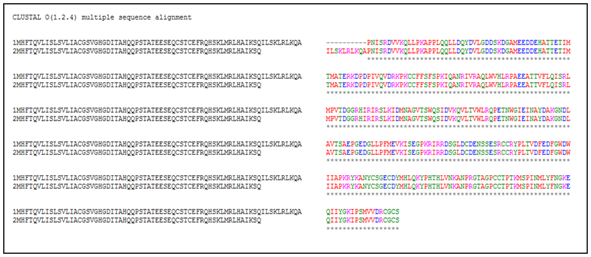
The retrieved sequence of myostatin from NCBI and UniProtKB heaving the accession number AEN75195.1 and entry no. G3EIK5 was subjected to multiple sequence alignment through Clustal Omega for finding out the sequence similarity. The multiple sequence alignment showed the almost close sequence similarity or identity. But, a little number of differences exists among the retrieved sequences of the different databases (Figure 1), which denotes that clustal Omega or clustal X is a useful web tool for finding out such differences. Therefore, it is required to consider any one database sequence before proceeding for further analysis during such studies. Moreover, the retrieved sequence of Myostatin from NCBI heaving the accession number AEN75195.1 was considered as the query sequence for further analysis.
Physicochemical properties of myostatin
The physicochemical properties of Myostatin were analyzed by Expasy protParam server. Myostatin is a 378 amino acid long protein with the estimated molecular weight 42613.74kDa. The theoretical isoelectric point (pI) is 6.42 and the amino acid composition showed the maximum presence of Leucine (7.7%) and minimum presence of Tryptophan (1.6%) (Table 1). The total number of negatively charged and positive charged residues of Myostatin are (Asp+ Glu)-48 and (Arg+ Lys)-45, respectively. The formula C1863H2959N527O565S26 of Myostatin contains 5940 atoms in total. The extinction coefficients of the concerned protein were calculated at 280 nm in water (M-1 cm-1) which presented the values of ext. coefficient 47285 and Abs 0.1% (=1g/l) 1.110, assuming all pairs of Cys residues form cystines; but the ext. coefficient becomes 46410 and Abs 0.1% (=1g/l) is 1.089 when assumed all Cys residues are reduced. The N-terminal of the sequence of the protein is Methionine (Met). The estimated half-life of Myostatin presented 30 hours in case of in-vitro culture condition of mammalian reticulocytes; whereas more than 20 hours for in-vivo culture condition of yeast and more than 10 hours in case of E. coli. The estimated instability index (ll) of the Myostatin is 54.27 which classify the protein as unstable. Aliphatic index 78.41 of the protein measures its considerable thermostability along with the relative volume occupied by aliphatic side chains. The value -0.430 of the GRAVY indicates that hydrophilic nature of the protein.
Amino Acids |
No’s. |
Percentage (%) |
Ala (A) |
22 |
5.80% |
Arg (R) |
22 |
5.80% |
Asn (N) |
12 |
3.20% |
Asp (D) |
26 |
6.90% |
Cys (C) |
14 |
3.70% |
Gln (Q) |
20 |
5.30% |
Glu (E) |
22 |
5.80% |
Gly (G) |
21 |
5.60% |
His (H) |
11 |
2.90% |
Ile (I) |
25 |
6.60% |
Leu (L) |
29 |
7.70% |
Lys (K) |
23 |
6.10% |
Met (M) |
12 |
3.20% |
Phe (F) |
10 |
2.60% |
Pro (P) |
23 |
6.10% |
Ser (S) |
27 |
7.10% |
Thr (T) |
22 |
5.80% |
Trp (W) |
6 |
1.60% |
Tyr (Y) |
9 |
2.40% |
Val (V) |
22 |
5.80% |
Table 1 Amino acid composition of Myostatin
Secondary structure prediction
The secondary structure of Myostatin predicted by PSIPRED server revealed the presence of helix region, strand region and coil regions (Figure 2). PSIPRED works on PSI-blast homology search algorithm.22 GOR IV analysis suggested the presence of alpha helix 23.54%, extended strands 21.16% and coiled region of 55.29%.
Template identification
For homology modeling, template 3hh2 (A) was identified showing 89% sequence similarity along with z-score of 987.78 from the PDBsum database (Figure 3). The selected template contains the crystal structure of myostatin: follistatin 288 complexes by X ray diffraction technique (2.15Å). The template sequence was also verified by SWISSMODEL/Workspace possessing the no. 3hh.2.1.A with 88.99% sequence identity with the query sequence (Figure 4).
Homology modeling
Homology modeling predicted the 3D structure of the Myostatin protein of Labeo calbasu. The structural conformation was generated from Swiss model server aligning the query sequence to the template sequence (Figure 5A & 5B). Model quality was estimated by assessing the QMEAN score, which stands for qualitative model energy analysis is composite scoring function describing the major geometrical aspects of protein structures. QMEAN was tested on several standard decoy sets including a molecular dynamics simulation decoy set as well as on a comprehensive data set. QMEAN shows a statistically significant improvement over nearly all quality measures describing the ability of the scoring function to identify the native structure and discriminate good from bad models.31
Validation of the prediction
The validation of the predicted structure (Figure 6A & 6B) of the proteins was done through Ramachandran plot (phi/psi). The stereochemical analysis of RAMPAGE server showed number of residues in favoured region is 96.3% and in allowed region is 3.7% (Figure 6), which indicates that predicted models is likely to be correct.
Protein model database
The generated model for myostatin was successfully submitted in the Protein Model Database (PMDB) heaving the PMID: PM0081037 (Figure 7).
Subcellular localization
By the application of Hum-mPLoc 2.0, extra cellular localization was obtained for the Myostatin protein.
The in-silico investigation of Myostatin revealed the physicochemical and structural parameters of the protein involved in prohibition or inhibition of skeletal muscle growth. Based on the findings, it could be concluded that Myostatin is an unstable, hydrophilic protein with secondary structure of alpha helix 23.54%, extended strands 21.16% and coiled region of 55.29%. The generated 3D structure of the Myostatin protein of Labeo calbasu by following homology modeling method was submitted to Protein Model Database (PMDB) which can be retrieved by the PMID no: PM0081037. Moreover, it might be resolved that the predicted 3D structure exhibited favoured region of 96.3% and allowed region of 3.7% which indicates that model is expected to be correct in prediction. The modeled structure of Myostatin can further utilized for predicting or designing blocker or inhibitor of the concerned protein. It can shed a new direction for the growth promotion of piscine species in a rapid manner in different developmental stages of life cycle. Further, extra cellular localization of Myostatin protein is also predicted by web tool which reduces the cost and effort during the experimental examination processes such as: immunohistochemistry, immunofluorescence etc. X-ray crystallography and NMR spectroscopy are most convenient method of 3D structure prediction, but requires enormous time and financial support, however also a tedious method in application. Moreover, the utility of computational and web tools minimizes these efforts. It is also useful for filling up the gaps generated within the large amount of data with available sequences and solved structures.
Authors are thankful to the Dean, College of Fisheries, Central Agricultural University (I), Lembucherra, Tripura (W) for encouragement and moral support. This work has been carried out under DBT funded project “Establishment of Bioinformatics Infrastructure Facility for Biology Teaching through Bioinformatics” and hence financial support by the Department of Biotechnology, Ministry of Science and Technology, Govt. of India, New Delhi is duly acknowledged.
The author declares no conflict of interest.

©2017 Acharjee, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.