International Journal of
eISSN: 2573-2889


Mini Review Volume 8 Issue 1
1Laboratory of Bioinformatics and Genomics of Microorganisms, Institute of Biological Sciences, Federal University of Para, Brazil
2Laboratory of Applied Genetics, Socio-environmental and Water Resources Institute, Federal Rural University of the Amazon, Brazil
3Institute of Animal Health and Production, Federal Rural University of the Amazon, Brazil
Correspondence: Marília Danyelle Nunes Rodrigues, Laboratory of Applied Genetics, Socio-environmental and Water Resources Institute, Federal Rural University of the Amazon, Belém, Brazil
Received: March 25, 2025 | Published: April 10, 2025
Citation: Macedo DB, Correa RMDS, Coelho HJP, et al. Mutations in GDF8 gene in double-muscled cattle breeds: an overview of mechanisms of action and its relevance to livestock production. Int J Mol Biol Open Access. 2025;8(1):20-25. DOI: 10.15406/ijmboa.2025.08.00193
Double muscling is a trait associated with mutations in the MSTN (GDF8) gene, which encodes myostatin, a protein belonging to the transforming growth factor beta (TGF-β) supercytokine family. Myostatin plays a crucial role in regulating muscle growth by maintaining satellite cells in a quiescent state and influencing cell development, proliferation, differentiation, migration, and apoptosis. Mutations in the GDF8 gene have been shown to be advantageous, as they confer important traits for beef cattle production, leading to strong selection for their use in breeding programs. The muscle hypertrophy-associated mutation is located in the MSTN (GDF8) gene. In certain double-muscled cattle breeds, different mutations result in the loss of myostatin function. A literature review was conducted on GDF8 gene mutations in various double-muscled cattle breeds, and the frequency of polymorphisms was investigated. Among the most frequently identified mutations were Indel c.818 and MSTN-F94L. Studies have evaluated the growth and reproductive traits of beef heifers to determine the impact of myostatin polymorphisms on reproductive performance, revealing that the MSTN Leu allele influences birth weight. Myostatin polymorphisms can be utilized to enhance carcass traits without compromising fertility in beef heifers. However, the overall effect of this genetic marker on herd performance remains uncertain. Therefore, before the beef industry adopts the MSTN-F94L marker for cattle selection, it is essential to fully understand its association with reproductive performance in cattle herds. Further studies are necessary to elucidate the molecular mechanisms underlying the effects of the MSTN-F94L polymorphism.
Keywords: double musculature, myostatin gene, SNP, genetical enhancement, beef cattle
Livestock farming is currently one of the most significant economic sectors in Brazil and globally. The restructuring of the beef production chain has become a priority for producers to meet the meat quality standards demanded by both domestic and international consumer markets.1 Increasing muscularity in cattle is widely regarded by researchers and producers as one of the primary objectives of genetic improvement programs. First described in the 19th century by Culley,2–4 the condition known as double musculature (DM) syndrome results from a mutation observed in various cattle breeds worldwide.5,6 This syndrome is characterized by muscle hypertrophy, particularly in the hindquarters, leading to a higher muscle-to-bone ratio and reduced fat deposition.7–9 The mutation in the GDF8 gene has been shown to be advantageous due to its significant attributes for beef cattle production, including improved feed conversion efficiency and meat with lower intramuscular fat content, characterized by a higher concentration of unsaturated fatty acids. These factors contribute to a healthier meat option for consumers. The zootechnical relevance of these traits has driven intensive selection for their incorporation into crossbreeding programs in Brazil and worldwide.10,11 Although cases of double musculature in cattle have been documented for over a century,12 the targeted selection for this trait only intensified after World War II. This was facilitated by advancements in veterinary medicine, particularly the use of anesthesia and antibiotics in assisted calving. Selection for double musculature can lead to dystocia due to morphological imbalances between the cow and the calf at birth. These imbalances may arise from an increase in calf width and weight in homozygous individuals, a reduction in the dam’s pelvic area, or a combination of both, depending on the mating strategy.13 However, heterozygous carriers of the double-muscling gene do not exhibit significant differences in calving ease compared to normal cattle, yet they demonstrate superior postpartum production traits.14 Further research is needed to elucidate the genetic relationships between double musculature and other production traits in beef cattle. In this context, the objective of this study was to review the literature on mutations in the GDF8 gene across different double-muscled cattle breeds. Understanding the role of this protein is essential for optimizing performance and meat production potential, as well as for refining breed selection strategies in crossbreeding and genomic selection programs.
Double muscling (DM) in cattle has been reported for over a century.12 There is a persistent trend to improve carcass quality in specialized beef breeds. As a result, a higher meat yield and more lean meat are desirable for the meat industry.15 Around the world, breeds that show productive and reproductive potential are selected for research. In some breeds of cattle with DM, it is possible to observe different mutations that lead to loss of function of the myostatin gene. The Belgian Blue breed, which originated in Belgium in the 1970s, is known for its extreme degree of muscularity, caused by a loss-of-function mutation in the myostatin gene (MSTN), also known as GDF8.16 The Aberdeen Angus breed stands out in the market today. This is due to the quality of the meat and its adaptation to Brazil's tropical climate. According to WEBER et al.17 the Angus breed had the highest number of semen doses sold in Brazil (16.4% of the total). This data shows the significant importance of this breed in the Brazilian meat production chain. Piedmontese cattle have high muscle mass and low fat deposition, low skeletal weight, low feed consumption and good feed conversion.14 Another extremely important breed for the beef market is the Marchigiana breed, which originated in Italy.18 The breed has a high proportion of muscle and a low proportion of fat and bone, which are considered economically desirable characteristics.19–21
Myostatin (MSTN), discovered in 1997 by Lee and McPherron,22 has since become a focal point of research aimed at understanding its function and potential inhibition for therapeutic applications, particularly in conditions associated with severe muscle mass loss. MSTN, a member of the transforming growth factor-beta (TGF-β) superfamily of cytokines, plays a critical role in regulating muscle growth by maintaining satellite cells in a quiescent state. It also influences various cellular processes, including development, proliferation, differentiation, adhesion, migration, and apoptosis.23 During the embryonic period of animal development, particularly during the process of myogenesis (Figure 1a), the GDF8 gene plays a critical role in regulating myoblast proliferation through its interaction with the Activin type IIB receptor (ActRIIB) (Figure 1b). This interaction inhibits somite activation by repressing the transcription factor Pax3, which subsequently impedes myoblast proliferation by down regulating MyoD and Myf5, and prevents myocyte differentiation into myotubes by suppressing the expression of myogenin and MRF4.24 Additionally, during embryogenesis, myostatin is expressed in cells of the myotome and developing skeletal muscle, where it regulates the final number of muscle fibers formed. In adulthood, myostatin continues to exert an inhibitory effect on skeletal muscle growth by suppressing key transcription factors involved in the muscle development cycle, thereby preventing muscle tissue hyperplasia.23
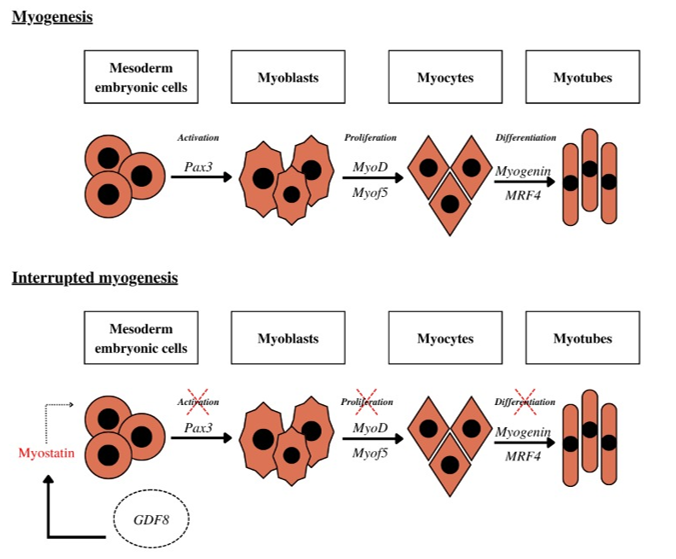
Figure 1 Stages of muscle tissue formation, or the myogenesis process, and mechanisms of gene action. (a) Steps in muscle tissue formation, or the myogenesis process, without interference from gene expression regulation mechanisms, (b) Stages of muscle tissue formation or the myogenesis process under the influence of myostatin and gene expression regulation mechanisms.
The muscle hypertrophy (mh) mutation is located in the MSTN gene, also known as growth and differentiation factor 8 (GDF8).25 In cattle, the MSTN gene has been mapped to the distal end of chromosome 2.16 The presence of the mutant mh allele in homozygous loss-of-function individuals leads to pronounced muscle hypertrophy. However, the mutation does not necessarily need to be present in the homozygous condition for its effects to be observed.26 Bos taurus was the first bovine species in which a mutation in the MSTN gene was identified. To date, 20 different mutations have been reported in the MSTN gene associated with double musculature, 14 of which are located in the coding sequence and six in the intronic regions.16,27–29 The first mutations in the GDF8 gene in cattle were described in two breeds, Belgian Blue and Piedmontese, both known for their significantly increased musculature compared to conventional cattle. Mutations in GDF8 have also been identified in various other species, including dogs,30 sheep,31 pigs,32 cattle,16 and even a documented case in a human.33
Over the past 12 years (2012–2024), several promising studies have explored the association between the GDF8 (MSTN) gene and mutations in various cattle breeds (Graph 1). However, no studies on MSTN gene polymorphism in cattle were found for the years 2012, 2016, 2017, 2019, and 2020. According to CASAS & KEHRLI,34 the MSTN gene significantly influences cattle traits, including the double-muscle phenotype and other economically important characteristics. Consequently, MSTN holds potential as a molecular marker in breeding programs.35 In the present study, we investigated the frequency of polymorphisms and identified Indel c.81836 and MSTN-F94L37 as the most commonly reported variants. The study analyzed single nucleotide polymorphisms (SNPs) and an 11 bp deletion in the coding region of the MSTN gene, assessing their relationship with the double-muscle phenotype in Belgian Blue crossbred cattle. Notably, four SNPs and an 11 bp deletion were identified within the MSTN coding region.36 The MSTN-F94L polymorphism has been shown to influence muscle and meat quality traits in a Jersey-Limousin backcross population.38,39 A study evaluating the impact of the F94L variant on genomic predictions using whole-genome SNP markers found that MSTN-F94L, located within the 6 Mb region of Bos taurus autosome 2 (BTA2), is strongly associated with birth weight, direct calving ease, maternal milk production, weaning weight, and carcass yield grade in both purebred Limousin and Limousin-Angus crossbred cattle.37 Furthermore, an analysis of Italian beef cattle breeds–Marchigiana, Chianina, Romagnola, Maremmana, and Podolica–identified a significant association between six SNPs in the BTA2 region (1.2–8.8 Mb) and muscularity in the Marchigiana breed. These SNPs include rs3423130174, rs43286831, rs109358737, rs43109236, rs110371799, and rs133461879.40
SNP records in the GDF8 gene associated with traits of zoo technical interest and their respective breeds of occurrence, reported between 2014 and 2024, are summarized in Table 1. Belgian Blue cattle are known for their exceptional carcass quality; however, they are also highly susceptible to psoroptic mange, a parasitic disease caused by the mite Psoroptes ovis. The study by MEYERMANS et al.5 identified an association between the myostatin gene mutation responsible for double musculature (mh/mh genotype) in Belgian Blue cattle and increased susceptibility to psoroptic mange. In this context, individuals exhibiting the double-muscle genotype and phenotype were more prone to developing psoroptic mange and presented with larger lesions. This association is relevant not only for other cattle breeds but also for other livestock species, as it suggests a negative correlation between double musculature and resistance to ectoparasites. In the Blonde d'Aquitaine breed, the TG3811 mutation in the second intron of the myostatin gene has been linked to superior carcass yield and muscle quality in homozygous [G/G] individuals. Additionally, cattle with the double-muscling genotype and phenotype exhibited higher carcass yield and reduced adiposity compared to heterozygous individuals. These findings suggest that this mutation likely has a direct effect on lipid metabolism, in addition to its well-documented impact on muscle fiber growth and differentiation.41 Furthermore, these characteristics have been associated with previously described mh mutations responsible for double musculature in cattle, such as nt821, C313Y, and Q204X, which have been identified in Limousin, Hereford, Piedmontese, Charolais, and Blonde d'Aquitaine breeds.42,43
|
Nº |
SNP |
Region |
Genotype |
Subspecies |
Breed |
Affected parameters |
References |
|
1 |
nt821(del1) |
11 bp, Exon 3 |
mh/mh |
Bos taurus taurus |
Belgian Blue |
Increased susceptibility to psoroptic mange and presence of double musculature |
Meyermans et al.5 |
|
2 |
T3811>G3811 |
41 bp, Intron 2 |
G/G |
Bos taurus taurus |
Blonde d'Aquitaine |
Highest musculature score, increased birth weight, higher dressing percentage, greater carcass weight, enhanced thigh width, and presence of double musculature |
Vinet et al.41 |
|
3 |
Indel c.818 (rs382669990) |
11 bp, Exon 3 |
del.11/del.11 g |
Bos taurus indicus |
Peranakan Ongole |
Increased birth weight and presence of double musculature |
Jakaria et al.44 |
|
4 |
Indel c.818 (rs382669990) |
11 bp, Exon 3 |
del.11/del.11 g |
B. taurus × B. indicus |
Belgian Blue x Peranakan Ongole |
Increased birth weight and presence of double musculature |
Jakaria et al.44 |
|
5 |
E291X |
874 bp, Exon 3 |
G/T |
Bos taurus taurus |
Marchigiana |
Increased slaughter weight, presence of double musculature, reduced fat content, and higher ash content |
Ceccobelli et al.45 |
|
6 |
g.-371T > A |
Regiao promotora |
A/T |
Bos taurus taurus |
Marchigiana |
Increased back width |
Sarti et al.19 |
|
7 |
g.874G>T |
874 bp, Exon 3 |
G/T |
Bos taurus taurus |
Marchigiana |
Increased croup and shoulder width |
Sarti et al.19 |
|
8 |
MSTN F94L |
94 bp, Exon 1 |
Leu:Leu |
Bos taurus taurus |
Crossbreed (1/8 Angus, 1/8 Hereford, 1/4 Braunvieh, 1/4 Limousin e 1/4 Charolais) |
Reduced birth weight and delayed onset of puberty |
Cushman et al.48 |
|
9 |
MSTN F94L (rs41638273) |
6 Mb, Cromossomo 2 |
- |
Bos taurus taurus |
Limousin; Crossbreed (1/2 Angus, 1/2 Limousin) |
Additive effect on birth weight; improved direct calving ease; increased ribeye area and yield grade |
Lee et al.37 |
|
10 |
MSTN F94L (rs110233897) |
6 Mb, Cromossomo 2 |
- |
Bos taurus taurus |
Limousin; Crossbred (1/2 Angus, 1/2 Limousin) |
Additive effect on birth weight; improved direct calving ease; increased ribeye area and yield grade |
Lee et al.37 |
|
11 |
MSTN F94L (rs109447543) |
38 Mb, Cromossomo 6 |
- |
Bos taurus taurus |
Limousin; Crossbreed (1/2 Angus, 1/2 Limousin) |
Additive effect on birth weight; improved direct calving ease; increased weaning and yearling weights |
Lee et al.37 |
|
12 |
MSTN F94L (rs110834363) |
38 Mb, Cromossomo 6 |
- |
Bos taurus taurus |
Limousin; Crossbreed (1/2 Angus, 1/2 Limousin) |
Additive effect on birth weight; improved direct calving ease; increased weaning and yearling weights |
Lee et al.37 |
Table 1 GDF8 (MSTN) gene SNPs associated with health and production traits in cattle breeds
In the agricultural industry, the identification of the double-muscling trait based on the myostatin (MSTN) gene is of great importance for crossbreeding programs. Studies on animals exhibiting double musculature have garnered significant attention from breeders and geneticists, particularly following scientific evidence linking the double-muscle phenotype to polymorphisms in the MSTN gene in certain cattle breeds.44 Mutations in the MSTN gene can lead to a substantial increase in skeletal muscle mass, resulting in the double-muscle phenotype. This discovery has sparked extensive discussions regarding the exploration and characterization of MSTN polymorphisms across various bovine breeds. Several single nucleotide polymorphisms (SNPs) have been identified in the coding region of the MSTN gene, including c.111G/C (rs523392653), c.267G/A (rs383271508), c.1077C/A (rs466598800), and c.1083T/C (rs211583837). Notably, the crossbred population (Belgian Blue × Peranakan Ongole) exhibited a higher degree of polymorphism than the purebred Peranakan Ongole population. The elevated genetic variation observed in both Peranakan Ongole cattle and their crossbred counterparts (Belgian Blue × Peranakan Ongole) highlights the genetic diversity within these populations.36
Previous studies have reported an 11 bp deletion in exon 3 of the MSTN gene, specifically an 11 bp deletion (5'-ATGAACACTCC-3'; rs382669990) in Belgian Blue cattle in Indonesia.16,25 Additionally, an 11 bp deletion at position c.818 (5'-ATGAACACTCC-3'; rs382669990) in exon 3 of the MSTN gene has been identified in Belgian Blue cattle, leading to a reduction in the coding region (1,117 bp). This deletion was not observed in Peranakan Ongole (PO) cattle; however, in Belgian Blue × PO crossbred cattle, a heterozygous 11 bp deletion (indel 11 bp) was detected.36 The Marchigiana breed, a specialized Italian beef cattle breed, is characterized by efficient weight gain, high carcass yield, and an optimal carcass fat percentage. A mutation in the MSTN gene, specifically a G-to-T transversion, has been identified in this breed. Homozygous [G/G] individuals exhibit the standard phenotype without double musculature, whereas [T/T] homozygotes present the double-muscle phenotype, which can sometimes lead to survival complications. Heterozygous [G/T] individuals, however, display pronounced muscularity without associated defects. In breeding practices, [T/T] homozygotes are typically excluded from reproduction, while heterozygotes are preferred as breeding stock. CECCOBELLI et al.45 reported that heterozygous bulls achieved, on average, a final live weight 17 kg higher than that of homozygous bulls at slaughter. The observed increase in average daily carcass gain suggests a direct effect of the MSTN gene mutation. Additionally, heterozygous bulls exhibited a lower incidence of bone in steak dissection, though this difference was not statistically significant. This finding aligns with previous reports indicating skeletal reduction in animals carrying MSTN mutations.14,20 Furthermore, steak samples from heterozygous animals exhibited a significantly higher proportion of muscle tissue (67.51%, p < 0.05) and a lower proportion of subcutaneous and intermuscular fat (6.62%, p < 0.01) compared to homozygous bulls (60.33% and 10.37%, respectively). These results are consistent with the muscle mass increase attributed to the loss of MSTN function in Marchigiana cattle, leading to muscle fiber hyperplasia, hypertrophy, and reduced subcutaneous and intermuscular fat deposition.46,47
CUSHMAN et al.48 evaluated the growth and reproductive characteristics of beef heifers over a three-year period to assess the influence of MSTN polymorphism on reproductive performance. The results indicated that the Leu allele of MSTN affected birth weight. However, the overall impact of this marker on herd performance has yet to be fully determined. LEE et al.37 identified the presence of the F94L variant of the MSTN gene in the 6 Mb region of BTA 2 in both purebred Limousin cattle and crossbreeds with other breeds, such as ½ Angus hybrids. This variant was positively associated with increased birth weight, improved direct calving ease, higher maternal milk production, and greater weaning weight. Furthermore, the study demonstrated that incorporating the MSTN F94L variant as either a random or fixed effect in genomic evaluations enhanced the accuracy of predictions compared to relying solely on SNP markers from the BovineSNP50 BeadChip, thereby improving the efficiency of genetic predictions. HAI et al.49 edited a mutation in the MSTN gene to induce loss of function and subsequent muscle hypertrophy. The MSTN -/- Luxi mutation was introduced in Luxi bulls using the CRISPR/Cas9 genome editing technique. Performance analyses were conducted on the F1 and F2 generations of crossbred cattle (Luxi × Simental) carrying the MSTN mutation to evaluate both its phenotypic effects and transmissibility. The F1 generation consisted of progeny from the cross between Luxi bulls carrying the MSTN -/- Luxi mutation and Simental cows lacking the muscle hypertrophy genotype (MSTN +/+). In this generation, two genotypes were identified: one with a 6 bp deletion (g.507del (6)) at position 507 and another with a 115 bp deletion (g.505del (115)) at position 505. The subsequent cross between F1 (Luxi × Simental) MSTN +/- cows and Luxi MSTN -/- bulls produced the F2 generation. While the F1 generation exhibited significant increases in body weight and enhanced hindquarter development, the F2 generation showed a greater increase in body weight index alone, along with a loss of Simental breed-specific coat color characteristics. RYAN et al.50 analyzed the association of 21 SNPs in the MSTN gene with traits related to calving difficulty, carcass fat content, carcass conformation, and carcass weight across 12 beef and dairy cattle breeds, including Angus, Aubrac, Blonde d'Aquitaine, Belgian Blue, Charolais, Friesian, Hereford, Holstein-Friesian, Limousin, Salers, Shorthorn, and Simental. Among all the variants examined, the nt821 polymorphism was most strongly associated with calving difficulty, particularly when the homozygous deletion was present in either the calf or the dam. The F94L, Q204X, and nt821 mutations were linked to improved carcass conformation and higher carcass weight. Additionally, the nt374_51, F94L, and E226X variants were associated with enhanced carcass quality while also contributing to increased direct calving ease.
In the context of animal genetic improvement, the principle of welfare conservation should be prioritized, with an emphasis on avoiding unnecessary suffering in animals subjected to selection and genetic editing. Although the principle of welfare conservation in genetically modified animals was introduced by philosopher Bernard Rollin in 1995, it remains insufficiently defined within the broader scope of animal genetic modifications, particularly when applied for non-scientific purposes. This concept is often referred to as the “Frankenstein Syndrome,” which asserts that “any animals that are genetically engineered for human use should be no worse off, in terms of suffering, after the new traits are introduced into the genome than the parent stock was prior to the insertion of the new genetic material”.51 In 2020, researcher Adam Shriver proposed a revision to the principle of welfare conservation, suggesting that certain genetic modifications can be beneficial for both humans and animals. His proposal aimed to promote transparency and maintain public trust in food producers. Shriver established that “any animals that are genetically modified through the use of genetic technology, for purposes other than research, should be no worse off, in terms of suffering, than the parent stock was prior to genetic alterations”.52 More broadly, genetic modifications should follow a framework of positive compensation, providing advantages for both humans and animals. Examples include engineering cattle to be resistant to mastitis53 and to naturally lack horns,54 thereby eliminating the need for painful management practices such as dehorning.
Myostatin polymorphism holds promise for enhancing carcass traits without adversely affecting fertility in beef heifers. However, its overall impact on herd performance remains unclear. Therefore, before integrating the MSTN F94L marker into cattle selection programs, it is essential to fully understand its association with reproductive performance. Mutations in the GDF8 gene, which encodes myostatin, are predominantly observed in taurine breeds. Nonetheless, breeds such as Peranakan Ongole and their crossbreeds may also carry GDF8 mutations linked to the double muscling phenotype. This highlights the need for further investigation into the presence and effects of GDF8 mutations in zebu breeds. Moreover, the mechanism of action of the MSTN F94L polymorphism remains largely unknown, underscoring the importance of continued research. Given that most genotypes associated with double muscling are found in taurine breeds, the introduction of these alleles into zebu populations–commonly raised in tropical regions–could facilitate the incorporation of the double-muscling trait into breeding programs, ultimately improving beef production efficiency.
None.
The authors declared that there are no conflicts of interest.

©2025 Macedo, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.