International Journal of
eISSN: 2573-2889


Research Article Volume 8 Issue 1
1Laboratory of Molecular Genetics and Microbiology, Instituto Nacional de Investigación y Desarrollo Pesquero, Argentina
2Computational Biology Institute, Department of Biostatistics and Bioinformatics, Milken Institute School of Public Health, The George Washington University, USA
Correspondence: Trucco María Inés, INIDEP, Paseo Victoria Ocampo Nº1, Mar del Plata, Buenos Aires, Argentina, Tel +54 9 223 4862580
Received: March 04, 2025 | Published: April 4, 2025
Citation: Andreoli G, Pérez-Losada M, Trucco MI. Identification of Squalus spp. in the Argentine-Uruguayan common fishing zone (ZCPAU) using genetic DNA barcoding. Int J Mol Biol Open Access. 2025;8(1):8-13. DOI: 10.15406/ijmboa.2025.08.00191
The Squalus genus is divided into three taxonomic categories that comprise various species, although their nomenclature is frequently inconsistent across different ichthyologists and taxonomies. In the South western Atlantic Ocean, there are three known species of spiny sharks, S. acanthias, S. cubensis, and S. mitsukurii, with discrepant classifications. During campaigns conducted by the INIDEP Chondrichthyan Fisheries Program, 27 Squalus specimens from the Argentine-Uruguayan Common Fishing Zone (ZCPAU) and Argentine coast were collected and morphologically identified onboard. We then coupled DNA COI Barcode technology with BLASTn and BOLD IDS tools to molecularly validate those taxonomic classifications. COI gene sequences were generated for those 27 specimens and compared to available reference sequences (FISHBOL database) from S. acanthias, S. cubensis, S. mitsukurii, S. suckleyi, S. blainville, and S. megalops from different oceanic regions. Genetic distances (K2P) and phylogenetic relationships (via neighbor-joining phylogenetic tree and Median-joining haplotype network) were also estimated for the COI data. A total of thirteen samples identified by the BOLD Identification System did not match the Squalus species identified using morphological evidence. Our analyses also indicated that S. mitsukurii, S. acanthias, and S. cubensis are three clearly distinct species, while S. lobularis is deemed equivalent to S. mitsukurii (K2P=0.35%). Our haplotype network also confirmed the existence of S. acanthias, S. cubensis, and S. mitsukurii in the studied area. The application of these molecular techniques and analyses have proved effective at accurately identifying Squalus species; hence we recommend their use to validate former morphology-based taxonomic identifications.
Keywords: squalidae, sharks, COI, south western atlantic, barcode
Squalidae is considered one of the most challenging shark groups to classify;1 it consists of two genera: Cirrhigaleus, which contains three species, and Squalus, which taxonomic composition is still debated due to significant challenges in species differentiation caused by its considerable morphological similarity.2 Squalus is usually subdivided in three distinct groups, although their taxonomic names can vary among experts. Bigelow and Schroeder3 were pioneers in describing these groups using morphological evidence; they studied specimens from the western Atlantic Ocean and categorized them into acanthias, blainville-fernandinus, and megalops or brevirostris-cubensis. Later, these researchers renamed these categories after their most ancient representative, resulting in S. acanthias, S. fernandinus, and S. megalops.4 Garrick5 proposed a revised naming system based on similar traits but with certain changes, identifying S. fernandinus (Molina, 1782) as a synonym for S. acanthias, thereby creating the groups: S. acanthias, S. blainville, and S. megalops - cubensis. Cadenat and Blache6 also identified three species groups, providing an identification key for the S. acanthias, S. blainville, and S. megalops groups. Compagno et al.7 and later researchers8,9 also recognized them as S. acanthias, S. mitsukurii, and S. megalops, which is the nomenclature currently in use. Nonetheless, the identification of these Squalus groups is still unclear and its taxonomy remains controversial. In the southwestern region of the Atlantic continental shelf situated between 34°S and 41°S, two spiny shark species from the Squalus genus were recorded: S. acanthias and S. blainville.10 The sightings of S. blainville along the shores of Buenos Aires and Uruguay are likely to correspond to S. cubensis, while those recorded in deeper waters of the shelf are probably of S. mitsukurii.11 Some researchers have even questioned the existence of S. cubensis in the region, suggesting that these sightings might actually be S. megalops.12 In a subsequent taxonomic review of chondrichthyans found in the southwestern Atlantic Ocean below 34°S, Menni and Lucifora13 indicated that there are three species of spiny dogfish: S. acanthias, S. cubensis, and S. mitsukurii (Figure 1).

Figure1 Squalus species present in the Southwest Atlantic, a- S. acanthias, b- S. mitsukurii and c- S. cubensis.16
The species S. cubensis is classified within the S. megalops group, whereas S. mitsukurii is linked to the S. mitsukurii group.8 Current taxonomic key guides developed to identify sharks of the genus Squalus from this area14,15 effectively assist in recognizing S. acanthias (skin with noticeable white spots, the spine of the first dorsal fin originates just behind the inner margin of the pectoral fins), but they pose challenges at distinguishing between S. mitsukurii and S. cubensis (both with smooth, spotless skin, the spine of the first dorsal fin originates just above the inner margin of the pectoral fins). Key features available for identification include the diagonal distance from the snout tip's center to the inner edge of the nostrils, which is longer in S. mitsukurii and shorter in S. cubensis, as well as the type of dermal denticles, tricuspidate in S. mitsukurii and unicuspidate in S. cubensis. However, these characteristics are difficult to evaluate during research expeditions or commercial trips.16 Consequently, utilizing molecular methods such as DNA barcoding of the mitochondrial gene for the enzyme cytochrome C oxidase subunit 1 (COI) can aid significantly in differentiating between Squalus species. The goal of this study was then to use DNA COI barcoding to molecularly identify Squalus specimens collected from the Argentine-Uruguayan Common Fishing Zone and the Argentine coast morphologically identified onboard as S. acanthias, S. cubensis, S. lobularis and S. mitsukurii. By doing this, we expect to resolve ongoing issues related to their taxonomic classification. Additionally, we will compare them to other Squalus species from different oceans and latitudes to determine their patterns of geographic distribution in their area of occurrence.
The Department of Molecular Genetics and Microbiology at INIDEP received muscle samples from 20 Squalus specimens from 5 fishing hauls of the vessel BIP Eduardo Holmberg during the EH03/13 campaign, as well as 7 samples caught during a single haul of the same ship during the EH02/16 research effort. The locations where these specimens were collected are within the Argentine platform and the Argentine-Uruguayan Common Fishing Zone (ZCPAU) (Figure 2). Onboard the vessel, the specimens were identified to the species level using morphological features.
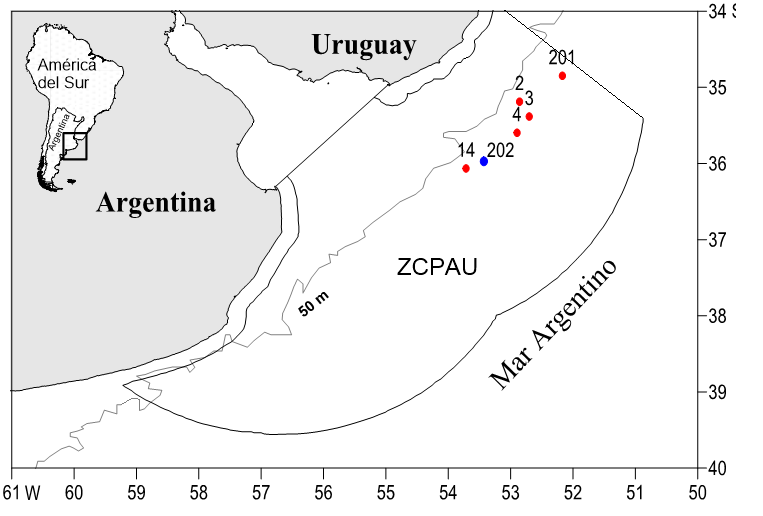
Figure2Location of the fishing hauls where the Squalus specimens under study were caught. Hauls from the EH03/13 campaign (20 specimens) and from the EH02/16 campaign (7 specimens) are indicated in red and blue, respectively
DNA extraction and PCR amplification
DNA was isolated from 50-100 mg of muscle tissue using phenol-chloroform-isoamyl alcohol; EB buffer (200mMl Tris pH 7.5, 250mM 5M NaCl, 25mM EDTA pH 8.0, 0.5% SDS) was used instead of CTAB in the DNA extraction, following Andreoli and Trucco.17 About 600 base pairs of the COI gene were amplified by PCR with the universal primers FishF2 (5' TCG ACT AAT CAT AAA GAT ATC GGC AC 3') and FishR1 (5' TAG ACT TCT GGG TGG CCA AAG AAT CA 3') as outlined by Ward et al.18 Every PCR reaction was performed in a total volume of 25 µl, which included: 1X PCR buffer (1.5 mM MgCl2), dNTPs (0.2 mM), primers (0.5 µM), 0.625 U of Taq polymerase (GoTaq DNA polymerase from Promega), and 2 µl of the DNA template. The amplification was conducted using a BioerLife Express TC-96/G/H thermal cycler with the following settings: an initial denaturation step of 2 min at 95º C; followed by 34 cycles consisting of 1 min at 94º C, 1 min at 54º C, and 2 min at 72º C; and a final extension of 15 min at 72º C. A negative control was used to ensure lack of contamination in all reactions. The integrity of the amplified products was evaluated through 1.5% agarose gel electrophoresis, with visualization accomplished using 0.01% ethidium bromide. The purified amplification products were then sequenced in both directions utilizing the Sanger technique at the Genomics Unit of INTA-Castelar, Argentina (http://www.inta.gov.ar).
Sequences analyses
Each sequence underwent manual review to identify uncalled or miscalled bases and all variable positions were verified by cross-referencing the sequence reads generated from both the forward and reverse strands of each individual. Manual editing of chromatograms and the assembly of both strands were carried out with the BIOEDIT v.5.0.6 program.19 The generated sequences were submitted to Genbank and assigned accession numbers OR707029 to OR707032 for S. acanthias, OR707039 to OR707043 for S. cubensis, and OR707044 to OR707055 for S. mitsukurii. The identification of each sequence was done through the Barcode of Life Data System (BOLD,20 using the Species Level Barcode Records option. Our COI sequences were combined with other COI reference sequences of S. acanthias, S. cubensis, S. mitsukurii, S. blainville, S. megalops, and S. suckleyi from the FISHBOL database, resulting in a final data set of 65 reference sequences. These species came from various locations, including the South and North Pacific, the Southwestern Atlantic, New Zealand, Italy, and the Gulf of Mexico (see Appendix). COI sequences were aligned using the Clustal W method and a model of evolution estimated. Phylogenetic relationships and genetic distances were inferred using the Neighbor-Joining (NJ) method under the best-fit model of evolution (K2P). Tree uncertainty was assessed using bootstrap analysis (5,000 replicates). An external outgroup sequence from the mandarin dogfish of the Squalidae family, Cirrhigaleus barbifer (Tanaka, 1912) (Genbank accession number 398719.1), was used to root the tree. Since our main goal was to identify Squalus species using COI barcodes and the BOLD methods, we did not consider necessary to use other more powerful maximum likelihood and Bayesian approaches of phylogenetic inference. All these analyses were performed in MEGA 6.21 The DnaSP v.5.1 package22 was also used to estimate haplotypes, variable sites, and haplotype diversity. Network haplotype relationships with their geographical distributions were inferred using the Median-Joining method23 in Network 10.1.24
Genetic identification of specimens obtained on board
The COI gene served as a useful molecular marker for recognizing specimens from the three Squalus species. The BOLD Identification System enabled species-level identification, achieving similarity percentages >99% for the best matches in every instance. Each specimen evaluated corresponded to the species S. acanthias, S. cubensis, or S. mitsukurii, which can be found in the area (Table 1). A total of thirteen samples identified by the BOLD Identification System did not match the Squalus species identified using morphological evidence. Five samples morphologically identified as S. cubensis were barcoded as S. acanthias (Sqlus 9, 13, and 16) and S. mitsukurii (Sqlus 5 and 6). Similarly, one sample morphologically identified as S. acanthias (Sqlus 11) was actually S. mitsukurii, and seven samples labeled as S. lobularis were confirmed to be S. mitsukurii (Table 1). These misclassifications involving S. acanthias, S. cubensis, and S. mitsukurii highlight the difficulty and ambiguity in using morphological features to distinguish these Squalus species effectively.
Phylogenetic relationships among Squalus species
Evolutionary relationships among selected Squalus samples were inferred using the NJ approach under the K2P model of evolution (Figure 3). The same model was used to estimate distances between Squalus species (Table 2). The NJ tree (Figure 3) depicted three distinct clades supported by bootstrap proportions (bp) of ≥99%. GROUP 1 contained sequences of S. acanthias and S. suckleyi grouped in two subclades (bp = 100%) and showed a K2P distance of 0.83%. The second clade (GROUP 2) encompassed sequences of S. blainville, S. megalops, and S. cubensis. Within this cluster, the sequences numbered 19, 2, 15, 18, 17, and 20 were placed alongside S. cubensis, affirming their classification. Furthermore, the tree depicted a clear distinction between S. cubensis and S. blainville or S. megalops (bp = 100%), also supported by K2P distances >1.50; this confirms S. cubensis is a different species from the other two. The third clade (GROUP 3) comprised S. mitsukurii and the specimens identified as S. lobularis (S. lob# in the tree). Both the NJ topology and the distance metrics indicated that the putative S. lobularis and S. mitsukurii are probably members of the same species. The distance between them was 0.35%, which overlaps with the intraspecific distances estimated for the other species (S. mitsukurii K2P=0.37 and S. lobularis K2P=0.33, data not shown).
|
S. acanthias |
S. suckleyi |
S. blainville |
S. megalops |
S. cubensis |
S. lobularis |
S. mitsukurii |
|
|
S. acanthias |
|||||||
|
S. suckleyi |
0.83 |
||||||
|
S. blainville |
8.08 |
7.90 |
|||||
|
S. megalops |
8.18 |
7.68 |
0.82 |
||||
|
S. cubensis |
7.78 |
7.30 |
1.73 |
1.53 |
|||
|
S. lobularis |
7.05 |
6.28 |
6.74 |
6.89 |
6.30 |
||
|
S. mitsukurii |
7.19 |
6.38 |
6.79 |
6.97 |
6.25 |
0.35 |
Table 2 K2P distances between species expressed as percentages
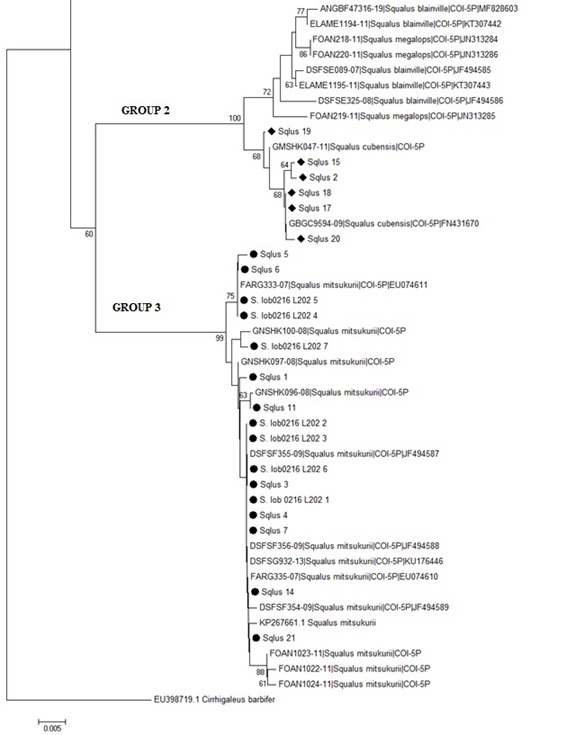
Figure3Neighbor joining tree of COI sequences from the Squalus species analyzed in this work (indicated with different symbols for each group) and those obtained from the FISHBOL database. Bootstrap proportions >50% are indicated
To study the relationships between the different Squalus species and their geographical distribution, a Median-Joining haplotype network was estimated (Figure 4). From the analysis of 92 sequences with 691 sites, 47 haplotypes were obtained, with a total of 86 variable sites, and a haplotype diversity (Hd) of 0.9247. Three distinct groups of Squalus were again obtained, GROUP 1 (for the acanthias-suckleyi group), GROUP 2 (for the megalops-blainville-cubensis group) and GROUP 3 (for mitsukurii-lobularis). Within each group the species showed a wide distribution, as evidenced by the diversity of colours in the haplotypes. The presence of the red colour, which corresponds to the South Atlantic, showed that it is possible to find species from all three groups (acanthias, cubensis and mitsukurii) in the ZCPAU and Mar Argentino. This analysis also confirmed that S. acanthias and S. suckleyi are distinct species. Although the number of mutational steps between the two species is low (three), a marked differentiation by distribution was observed. The haplotypes of the S. suckleyi sequences are not shared with those of S. acanthias and were limited to the North Pacific (the only haplotypes in black), while the haplotypes of S. acanthias had a wider distribution, from the South Atlantic (red) to the North Atlantic (green), South Pacific (grey), Italy (blue) and New Zealand (yellow). For group B, the species S. blainville and S. megalops were limited to Italy (blue) and South Africa (violet) and no shared haplotype was observed (i.e., two different species). S. cubensis occurred between the North (green) and South Atlantic Ocean (red), confirming its presence in our region and its separation from the other Squalus species, as also suggested by the NJ tree. Concerning S. mitsukurii and S. lobularis, our network also showed that they are actually the same species, not differentiated by their geographical locations, as they share identical haplotypes (in the predominant H1, sequences from both kinds are present). The sequences originating from Taiwan and southern China (orange haplotypes) are likely to be included in this group. The outlined distribution pattern is presented in Table 3.
|
Squalus species |
Distribution |
|
S. acanthias |
North Atlantic Ocean |
|
South Atlantic Ocean |
|
|
Mediterranean |
|
|
New Zealand |
|
|
South Pacific |
|
|
S. sukleyi |
North Pacific |
|
S. megalops y S. blainville |
Mediterranean |
|
South Africa |
|
|
S. cubensis |
North Atlantic |
|
South Atlantic |
|
|
S. mitsukurii-S.lobularis |
North Atlantic |
|
South Atlantic |
|
|
South Africa |
|
|
Taiwan |
Table 3 Distribution pattern of the Squalus species analyzed in this study
This study shows that DNA barcoding of COI sequences can be applied to identify and classify the occurrence of Squalus species in the southwestern Atlantic. This molecular approach is more accurate than using morphological evidence since external physical traits (the shape of dorsal, pectoral, and anal fins, color patterns and the dimensions of the dermal denticles) often overlap among various species leading to Squalus misidentification. The variation in classification between S. lobularis and S. mitsukurii underscores the complicated taxonomic status of these species, stemming from the categorization suggested by Viana et al.25 during their comprehensive global review of Squalus. These researchers conducted a regional reassessment of specimens from Brazil, Uruguay, and Argentina to determine the valid species in the southwest Atlantic Ocean. By carrying out an in-depth comparative study of the external features and skeletal structures of Squalus specimens in the region, they proposed that S. mitsukurii and S. cubensis were absent. Our findings contradict that conclusion since the genetic distance estimated between the specimens identified morphologically as S. lobularis and S. mitsukurii suggested that they actually are the same species (K2P= 0.35%). This was further validated by the observation of that both species shared haplotypes and occupied the same geographic regions, indicating that S. mitsukurii is present in the southwestern Atlantic. It is crucial to note that the research conducted by Viana et al.25 lacked molecular analysis and relied exclusively on morphological features.
Ariza et al.26 adopted the nomenclature of S. albicaudus for the Atlantic specimens from the Brazilian coast, following Viana et al.25 Their study covered specimens from the southwestern Atlantic and the Pacific. The K2P distance found by these authors between S. albicaudus and S. cubensis was 0.0072, so they speculated that S. albicaudus might encompass a population of S. cubensis in the Southeastern Atlantic Ocean that is presently experiencing speciation – such process would be relatively new and not entirely finished, given that these species still share haplotypes. The distribution to the southern region of Brazil is unknown.25 The DNA Barcode method applied in this study also allowed us to distinguish between S. acanthias and S. sukleyi, which corroborated previous findings by Ebert et al.9 regarding the differentiation between these two species using morphological, meristic and molecular data. The distance between both species analyzed in this study was 3 to 7 times greater than their respective intraspecific distances, which also agreed well with the distances in Ebert et al., who reported an interspecific distance up to 5 or 6 times greater. S. acanthias can be easily differentiated from the other two species found in the area due to the presence of white spots on both sides; however, distinguishing between S. mitsukurii and S. cubensis is more complicated. The main differences between these two species lie in the length of the prenarial region and the specific type of dermal denticles.14,15 These features are challenging to identify during research expeditions because Squalus sharks in the area often gather in groups, with some sets yielding up to 900 kg of S. mitsukurii. This high volume per set complicates the detailed analysis of individual specimens.16 This may explain why two male specimens (Sqlus 5 and 6) were initially identified as S. cubensis aboard and later identified through BOLD as S. mitsukurii (100% similarity). Additionally, six out of the eleven specimens labeled as S. cubensis showed 100% or nearly identical similarity to S. cubensis sequences from the Gulf of Mexico, which contrasts with the findings of Viana et al.25 regarding this species' limited distribution in the southwestern Atlantic. Accurate identification of species is crucial for implementing conservation strategies and ensuring the responsible use of natural resources. The species studied in this work are considered by the International Union for Conservation of Nature (IUCN) as vulnerable (S. acanthias), endangered (S. mitsukurii)27 and least concern (S. cubensis).28 In 2006, the "Spiny Shark" item was added to the official fishing statistics of Argentina. It appears as Dogfish sharks nei, so landings reported under this category may include any of the three species considered here.29 As Colonello et al.16 observed, it is important to enhance the identification of observable traits that can be quickly and easily recognized in S. mitsukurii and S. cubensis samples. This will facilitate research on the biology, habitat preferences, and stock dynamics of populations and species of Squalus in the area.
The current research established that S. cubensis and S. mitsukurii can be accurately identified through DNA COI barcode analysis. It also showed that the two species form distinct evolutionary lineages, as indicated by our NJ phylogeny, K2P distances and Median Joining haplotype network. Moreover, the network analysis confirmed the presence of S. mitsukurii, S. acanthias, and S. cubensis in the Argentine Sea and the Argentine-Uruguayan common fishing area (ZCPAU). Therefore, since these species cannot be easily distinguished aboard vessels, we recommend the use of the molecular methods employed in this study to validate initial morphological assessments and reach accurate taxonomic identifications.
The authors would like to thank Dr. Jorge Colonello for providing us with data, records and images useful for the development of this work, as well as the observers on board the research vessels. We also thank Silvina Izzo for her help in the analysis of sequences.
The authors declared that there are no conflicts of interest.

©2025 Andreoli, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.