
Research Article Volume 4 Issue 5
On estimating flexible weibull parameters with type I progressive interval censoring with random removal using data of cancerous tumors in blood
Afify WM
Regret for the inconvenience: we are taking measures to prevent fraudulent form submissions by extractors and page crawlers. Please type the correct Captcha word to see email ID.

Department of Head of Statistics, Mathematics & Insurance, Kafr El-sheikh University, Egypt
Correspondence: Afify WM, Department of Head of Statistics, Mathematics & Insurance, Kafr El-sheikh University, Faculty of Commerce, Egypt
Received: September 09, 2016 | Published: October 14, 2016
Citation: Afify WM. On estimating flexible weibull parameters with type I progressive interval censoring with random removal using data of cancerous tumors in blood. Biom Biostat Int J. 2016;4(5):208-216. DOI: 10.15406/bbij.2016.04.00108
Download PDF
Abstract
In this paper, the maximum likelihood and the Bayes estimators of the two unknown parameters of the flexible Weibull distribution have been obtained for progressive Interval type-I censoring scheme with binomial random removal. Point estimation and confidence intervals based on maximum likelihood and bootstrap method are also proposed. A Bayesian approach using Markov chain Monte Carlo (MCMC) method to generate from the posterior distributions and in turn computing the Bayes estimators are developed. To illustrate the proposed methods will discuss an example with the real data. Finally, comparing the two techniques through comparisons between the maximum likelihood using bootstrap method and different Bayes estimators using MCMC study.
Keywords: flexible weibull distribution, progressive interval type-I censoring, random removal, percentile bootstrap, bayesian and non-bayesian approach, markov chain monte carlo (MCMC)
Introduction
Censoring is very common in life tests in the past several decades; the experimenter may be unable to obtain complete information on failure times of all experimental items. For this reason, Aggarwalla1 suggested a useful type of censoring, namely, a progressively Type I interval censored data, which is a union of Type I interval and progressive censoring. This method of lifetime data collection can be useful to a biological experimenter, particularly when the experimental units are humans, as continuous monitoring is often not possible to implement, and withdrawal rates from such studies may high.
In progressive censored the number of units being removed from the test at each failure time may occur at random. For example; the number of patients who drop out of clinical test at each stage is random and cannot be predetermined. That is why to display a more general censoring scheme called progressive progressively Type I interval censored with random removal. It can be described as follows: suppose
units are put on life test at time
and under inspection at m pre-specified times
where
is scheduled time to terminate the experiment. The number,
, of failures within
is recorded and
surviving items are randomly removed from the life testing at the ith inspection time,
, for
. Since the number,
, of surviving items is a random variable and exact number of items with drawn should not be greater than
at time schedule
,
are random. Such a censoring mechanism is termed as progressive interval type-I censoring with random removal scheme. If we assume that probability of removal of a unit at every stage is π for each unit then ri can be considered to follow a binomial distribution i.e,
for
. The main difference between progressive interval type I censoring with fixed removal and progressive interval type I censoring with random removals is that the removals are predetermined in the former case while they are random in the latter case. Note that m is pre-determined in both cases. However, many practical applications suggest that it is more flexible to have removals random to accommodate the unexpected drop out of experimental subjects.
Although progressive censoring occurs frequently in many applications, there are relatively few works on it. Some early works can be found in Cohen,2 Readers can refer to the book Balakrishnan & Aggarwala3 for more details on the methods and applications of this topic. However, all these works assumed that the number of units being removed from the test is fixed in advance. In practice, it is impossible to pre-determine the removal pattern. Thus, Yuen & Tse4 and Yang et al.5 considered the estimation problem when lifetimes collected under a Type II progressive censoring with random removals and Kendell & Anderson6 point out that the expected duration under grouped data. Progressive type-I interval censored sampling is an important practical problem that has received considerable attention in the past several years. Based on the progressive type-I interval censored sampling, Ashour & Afify7 derived the maximum likelihood estimators of parameters of the exponentiated Weibull family and their asymptotic variances under random removal. Lin et al.8 determined optimally spaced inspection times for the log-normal distribution, while Ng & Wang9 and Chen & Lio10 compared three classical estimation methods, the maximum likelihood estimators the moment method and the probability plot method in terms of the Weibull distribution and generalized exponential respectively.
In Bayesian approach, It is too difficult to find integrate over the posterior distribution and the problem is that the integrals are usually impossible to evaluate analytically. But in MCMC technique, the MCMC methodology provided a convenient and efficient way to sample from complex, high-dimensional statistical distributions. Recently, application of the MCMC method to the estimation of parameters or some other vital properties about statistical models is very common. Green et al.11 using the MCMC method for estimating the three parameters Weibull distribution, and they showed that the MCMC method is better than the ML method, when given a proper prior distribution of the parameters. As a generalization of the two parameter Weibull model, Gupta et al.12 gave a complete Bayesian analysis of the Weibull extension model using MCMC simulation and complete sample. Lin & Lio13 discussed Bayesian inference under progressive type I interval censoring by using MCMC.
A random variable x is said to have a Flexible Weibull Distribution with parameters
if its probability density function, cumulative function, survival function and hazard function are given by
(1)
(2)
(3) respectively.
In this paper we consider the Bayesian inference of the scale parameters for progressive interval type-I censored data when both parameters are unknown. We assumed that the both scale parameters
have gamma prior and they are independently distributed. As expected in this case also, the Bayes estimates cannot be obtained in closed form. We propose to use the Gibbs sampling procedure to generate MCMC samples, and then using the Metropolis–Hastings algorithms, we obtain the Bayes estimates of the unknown parameters. We perform some simulation experiments to see the behavior of the proposed Bayes estimators and compare their performances with the maximum likelihood estimators.
The rest of the paper is organized as follows. In the next section, the ML estimators of the unknown parameters and approximate confidence intervals are presented. The corresponding parametric bootstrap confidence intervals for the parameters are given in Section 3. In Section 4, we cover Bayes estimates and construction of credible intervals using the MCMC techniques. In Section 5, for illustrative purposes, we performed a real data analysis. Comparisons among estimators are investigated through Monte Carlo simulations in Section 6. Finally, conclusions appear in Section 7.
Classical estimation and percentile bootstrap algorithm (Boot-p)
Classical estimation (maximum likelihood estimators) of the unknown parameters and approximate confidence intervals are presented. Also, the corresponding parametric bootstrap confidence intervals using percentile bootstrap Algorithm (Boot-p) for the parameters are given in this section.
Classical estimation
Suppose a progressively Type-I interval censored sample is collected as described above, beginning with a random sample of units with a continuous lifetime distribution
and let
denote the number of units known to have failed in the intervals
, respectively. Then, based on this observed data, the joint likelihood function will be Aggarwala.1
(4)
Where C is constant. Clearly, if
for
and
equation (4) reduces to the likelihood function for interval type I censoring data is defined as follows:
Where
and
are the number of units known to have failed in the intervals
respectively.
For type I progressive Interval censoring, supposed that
is independent of
for all
; Wu & Chang14 suggested the following likelihood function of a progressive interval censoring with binomial removals
(5)
Where
is the likelihood function for a progressive type I interval censored with fixed removal (4) and
will be
Such as
(6)
and
are the same as defined before in (1) and (2) respectively. The log likelihood function with random removal can be written as
(7)
The maximum likelihood estimations of
and
are the simultaneous solutions of following normal equations
(8)
(9)
Note that
does not involve the parameters. Therefore, the MLE
of
can be found by maximizing
directly, that is,
Therefore, the maximum likelihood estimation of parameter
is given by
(10)
It may be noted that (9) and (10) cannot be solved simultaneously to provide a nicely closed form for the estimators. Therefore, we propose to use fixed point iteration method for solving these equations. Using Fisher information matrix
in the Appendix and the asymptotic normality of the maximum likelihood estimators can be used to compute the approximate confidence intervals (ACI) for parameters
,
and
Therefore,
confidence intervals for parameters
,
and
will be become
,
and
Where
is percentile of the standard normal distribution with right-tail probability
.
Data algorithm
The data generation is based on the algorithm proposed by Aggarwala1 to simulate the numbers,
of failed items in each subinterval
from an initial sample of size putting on life testing at time 0. This algorithm, which is an extension from the procedure developed by Kemp & Kemp15 for the multinomial distribution, involves generating m binomial random variables. A procedure to generate a progressively type I interval censored data with random removal,
from the flexible Weibull distribution can be described as follows briefly: let
and
and for
Step 1 set
and let
.
Step 2
Using initial
to generate a sample
,
using binomial distribution, where
following the binomial
distribution and the variables
follow the binomial
distribution for
Set
Generate k_i as a binomial random variable with parameters n-k sum-r sum and
Step 3 Set
and
.
Step 4 If
, go to step 2; otherwise, stop.
Percentile bootstrap algorithm (Boot-p)
We can increase information about the population value more than does a point estimate by using a parametric bootstrap interval. We propose to use confidence intervals based on the parameteric bootstrap methods using percentile bootstrap Algorithm (Boot-p) based on the idea of Efron.16
The algorithm for estimating the confidence intervals is illustrated as follows:
Before progressing further, we first describe how we generate progressively interval Type I censored data with binomial random removals. The following algorithm is followed to obtain these samples.
- Specify the values of
.
- Specify the values of
and
.
- Form data algorithm; compute the maximum likelihood estimates of the parameters
,
and
, by solving the likelihood equations simultaneously in (8), (9) and (10).
- Use
,
and
, to generate a bootstrap sample
with the same values of r_i, m;(i=1,2,…,m) using algorithm presented in Balakrishnan & Sandhu.17
- As in step 3, based on
compute the bootstrap sample estimates of
,
and
, say
,
and
.
- Repeat steps 4-5 B times representing B bootstrap maximum likelihood estimators of
,
and
based on B different bootstrap samples.
- Arrange all
,
and
, in an ascending order to obtain the bootstrap sample
(where
,
and
).
Let
be the cumulative distribution function of
. Define
for given Z. The approximate bootstrap
confidence interval (ABCI) of
is given by
.
Bayesian estimation and MCMC technique
In this section, we will focus to Bayesian approach using Markov chain Monte Carlo (MCMC) method to generate from the posterior distributions and in turn computing the Bayes estimators are developed.
Bayesian estimation
In Bayesian scenario, we need to assume the prior distribution of the unknown model parameters to take into account uncertainty of the parameters. The informative prior densities for
and
are given as
,
,
and
has a
Note that the parameters
,
and
behave as independent random variables. The joint informative prior probability density function of
,
and
is
(11)
where
are assumed to be known and are chosen to reflect prior knowledge about
,
and .
Note that when
, (we call it prior 0) they are the non-informative
,
and
respectively.
It follows from (4), (6) and (11) that the joint posterior density function of
,
and
given x is thus
(12)
where
and
.
It is not possible to compute (12) analytically. The problem is that the integrals in (12) are usually impossible to evaluate analytically, and the numerical methods may fail. The MCMC method provides an alternative method for parameter estimation. In the following subsections, we propose using the MCMC technique to obtain Bayes estimates of the unknown parameters and construct the corresponding credible intervals.
MCMC technique
Computer simulation of Markov chains in the space of parameter will depend on Markov chain Monte Carlo (MCMC) Gilks et al.18 The Markov chains are defined in such a way that the posterior distribution in the given statistical inference problem is the asymptotic distribution. However, the posterior likelihood usually does not have a closed form for a given progressively type-I interval-censored data. Moreover, a numerical integration cannot be easily applied in this situation. A lot of standard approaches to display like Markov chains exist, including Gibbs sampling, Metropolis-Hastings (M-H) and reversible jump. The M-H algorithm is a very general MCMC method first expansion by Metropolis et al.19 and later extended by Hastings.20 it is possible to use these algorithms by implement posterior simulation in essentially any problem which allow point wise evaluation of the prior distribution and likelihood function. It can be used to obtain random samples from any arbitrarily complicated target distribution of any dimension that is known up to a normalizing constant. In fact, Gibbs sampler is just a special case of the M-H algorithm.
In order to use the method of MCMC for estimating the parameters of the flexible Weibull distribution and random removal, namely,
,
and
. Let us consider independent priors as in (10), the full conditional distribution for any parameter can be obtained, to within a constant, by factoring out from the likelihood function
any terms containing the relevant parameter and multiplying by its prior. From (11), the full posterior conditional distribution for
is proportional to
(12)
Also, the full posterior conditional distribution for β is proportional to
(13)
Similarly, the marginal posterior density of
is proportional to
(14)
It is noted that the posterior distribution of
is beta with parameters
and
where
and
and,
therefore, samples of can be easily generated using any beta generating routine. But the conditional posterior distribution of
and
equations (12) and (13) respectively, cannot be reduced analytically to well-known distributions and therefore it is not possible to sample directly by standard methods, but the plot of it show that it is similar to normal distribution. So to generate random numbers from this distribution, we use the M-H method with normal proposal distribution.
MCMC process
Now, we propose the following scheme to generate
,
and
from density functions and in turn obtain the Bayes estimates and the corresponding credible intervals.
- Start with an
and
.
- Set
.
- Generate
from beta distribution
.
- Using M-H algorithm Metropolis et al. [19],
from
with the
proposal distribution where
is the variance of obtained using variance-covariance matrix; similarly,
from
with the
proposal distribution where
is the variance of
obtained using variance-covariance matrix.
- Compute
,
and
.
- Set
.
- Repeats Steps 3-6 N times.
- Obtain the Bayes estimates of
,
and
with respect to the squared error loss function as
,
and
- To compute the credible intervals of , and , order
,
and
as
,
and
.Then the
symmetric credible intervals (SCI) of , and become:
and
Real data analysis
To conduct a study within the Institute of Oncology in Tanta - Egypt. This study is concerned with the treatment of cancerous tumors in blood and studies their impact on the overall health of the patient. Underwent the study 228 patients and they had varying degrees of disease. Patients were examined every 15 days for 6 consecutive months. Of course there were cases of withdrawal (death - interruption of treatment for different reasons)
As we know on the basis of a single sample, one cannot make a general statement regarding the behavior of proposed estimators, therefore we present a simulation study for the study of the behavior of the estimators in the next section.
Simulation
The simulation is conducted using the R version 3.2.2 (for more information about R programming, the reader may refer to this manual of R, version 3.3.0 under development (2015-10-30) Copyright 2000 –2015 R Core Team). The simulation setup is parallel to the real data given in (Table 1). To be specific, each replication of the simulation generates a progressively type-I interval-censored data within twelve subintervals which have pre-specified inspection times (in terms of half month),
The last inspection time, , is the scheduled time to terminate the experiment. The lifetime distribution is flexible Weibull with parameters
and
where the simulation input parameters are selected close to the maximum likelihood estimators of flexible Weibull parameters for modeling the real data in (
Table 1). The performance of parameter estimation under progressively type I interval censored with random removal is compared via the maximum likelihood, bootstrap method and MCMC procedure developed in this paper. The summary for 1000 simulation runs is shown in (
Tables 2-5).
Bayes estimates of
,
and
using MCMC method, we assume that informative priors a = 2,b = 3,c = 4 , d = 2, A = 2 and B = 3) on
,
and
in (
Table 4). Also, by non-informative prior using MCMC procedure with Bayes estimation will be obtained on estimates of parameters in (
Table 5).
|
Cases of Withdrawal |
Number of Random Removals
|
Interval in Hours
|
Number at Risk |
Number of Failure
|
1 |
[0,16) |
228 |
25 |
2 |
2 |
[16,31) |
201 |
39 |
2 |
3 |
[31,46) |
160 |
25 |
1 |
4 |
[46,61) |
134 |
20 |
3 |
5 |
[61,76) |
111 |
11 |
1 |
6 |
[76,91) |
99 |
14 |
2 |
7 |
[91,106) |
83 |
11 |
3 |
8 |
[106,121) |
69 |
17 |
0 |
9 |
[121,136) |
52 |
6 |
2 |
10 |
[136,151) |
44 |
31 |
1 |
11 |
[151,166) |
12 |
6 |
1 |
12 |
[166,181) |
5 |
5 |
0 |
Table 1 Examine patients every 15 days
|
Different Parameters |
|
|
|
Average |
6.441 |
0.0841 |
0.6312 |
MSE |
0.0134 |
0.0743 |
0.0484 |
Bias |
0.0231 |
0.1073 |
0.0094 |
Variance |
0.0129 |
0.0627 |
0.0483 |
ACI |
[5.0132,7.9801] |
[-0.1736,0.0901] |
[0.4421,0.7782] |
Length ACI |
2.9669 |
0.2637 |
0.3361 |
Table 2 Progressively type I interval censored with random removal via the, maximum likelihood
|
Different Parameters |
|
|
|
Average |
5.966 |
0.099 |
0.7058 |
MSE |
0.1174 |
0.0984 |
0.0487 |
Bias |
0.0346 |
0.1764 |
0.0109 |
Variance |
0.1162 |
0.0673 |
0.0486 |
ABCI |
[5.0117,8.0412] |
[-0.1811,0.0884] |
[0.4434,0.7992] |
Length ABCI |
3.0295 |
0.2695 |
0.3558 |
Table 3 Progressively type I interval censored with random removal via the, bootstrap method
|
Different Parameters |
|
|
|
Average |
4.902 |
0.0083 |
0.5118 |
MSE |
0.0035 |
0.0277 |
0.04804 |
Bias |
0.0049 |
0.0833 |
0.0017 |
Variance |
0.0035 |
0.0207 |
0.04803 |
SCI |
[5.1023,7.6421] |
[-0.1075,0.0826] |
[0.4927,0.6524] |
Length SCI |
2.5398 |
0.1901 |
0.1597 |
Table 4 Progressively type I interval censored with random removal via the, MCMC procedure developed (Informative Priors)
|
Different Parameters |
|
|
|
Average |
4.671 |
0.0398 |
0.6501 |
MSE |
0.0673 |
0.0559 |
0.0656 |
Bias |
0.0174 |
0.0304 |
0.0093 |
Variance |
0.067 |
0.0549 |
0.0655 |
SCI |
[4.8821,8.0307] |
[-0.1010,0.0721] |
[0.3881,0.7061] |
Length SCI |
3.1486 |
0.1731 |
0.318 |
Table 5 Progressively type I interval censored with random removal via the, MCMC procedure developed (Non-Informative Priors)
Both of density functions of
and
can be approximated by normal distribution functions but density function of
will be beta as mentioned in subsection (3.3) which are plotted in (Figure 1& 2) Chain of MCMC outputs of
,
and
, using 100 000 MCMC samples. This was done with 1000 bootstrap sample and 100 000 MCMC sample and discard the first 50000 values as ‘burn-in’. The Bayes estimators can be seen to have the smaller risks than classical estimators for all the considered cases. It may also be noted that the Bayes estimators obtained under informative prior are more efficient than those obtained under non-informative priors. This indicates that the Bayesian procedure with accurate prior information provides more precise estimates. Also, The Length of the SCI (using informative prior) is smaller than the Length of the ACI and ABCI.
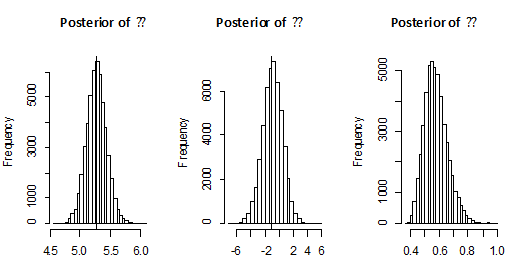
Figure 1 Posterior density function of
,
and
.
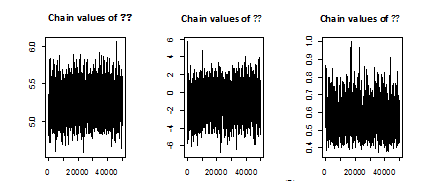
Figure 2 Chain of MCMC outputs of
,
and
.
Conclusion
The methodology developed in this paper will be very useful to the researchers, engineers, statisticians and in the field of medical where such type of life test is needed and especially where the Weibull distribution is used. we have considered the problem of estimation for flexible Weibull distribution in the presence of Progressive Type-I Interval censored sample with Binomial removals. The scope of this censoring scheme in clinical trials has been discussed. We have found that Bayesian procedure provides estimates of the unknown parameters of flexible Weibull model with smaller MSE. The length of SCI is smaller than that of the ACI and ABCI. Applying the MCMC process through the application of the MH algorithm to deal with the Bayesian estimation for another lifetime distributions under type I progressive interval censoring with random removal could be a fruitful future research.
Appendix
The asymptotic variance-covariance matrix of the maximum likelihood estimators for parameters, and are given by elements of the inverse of the Fisher information matrix with random removal will be
,
Unfortunately, the exact mathematical expressions for the above expectations are very difficult to obtain. Therefore, we give the approximate (observed) asymptotic varaince-covariance matrix for the maximum likelihood estimators, which is obtained by dropping the expectation operator E, where
We explained how to find Fisher information matrix
in the Appendix B.
,
and
Numerical technique is needed to obtain the Fisher information matrix and the variance-covariance matrix. Note that under fixed and random removal the estimates based on intervals with equal length when the intervals are of equal length, so that monitoring and censoring occur periodically say
.
We determine the second partials by differentiating the first partials, equations (8) and (9), obtaining
Note that:
Acknowledgments
Conflicts of interest
Author declares that there are no conflicts of interest.
References
- Aggarwala R. Progressive interval censoring: Some mathematical results with applications to inference. Communications in Statistics - Theory and Methods. 2001;30(8-9):1921‒1935.
- Cohen AC. Progressively censored samples in life testing. Technometrics. 1963;(3):327‒339.
- Balakrishnan N, Aggarwala R. Progressive Censoring: Theory, Methods, and Applications, Boston, MA: Birkh¨auser Boston. 2000.
- Yuen HK, Tse SK. Parameters estimation for Weibull distributed lifetimes under progressive censoring with random removals. Journal of Statistical Computation and Simulation. 1996;55(1-2):57‒71.
- Yang C, Tse SK, Yuen HK. Statistical analysis of Weibull distributed life time data under type II progressive censoring with binomial removals. Journal of Applied Statistics. 2000;27(8):1033‒1043.
- Kendell PJ, Anderson RL. An Estimation Problem in Life Testing. Technometrics. 1971;13(2):289‒301.
- Ashour SK, Afify WM. Statistical analysis of exponentiated Weibull family under type-i progressive interval censoring with random removals. Journal of Applied Sciences Research. 2007;3(12):1851‒1863.
- Lin CT, Wu SJS, Balakrishnan N. Planning life tests with progressively type-I interval censored data from the lognormal distribution. Journal of Statistical Planning and Inference. 2009;139(1):54‒61.
- Ng HKT, Wang Z. Statistical estimation for the parameters of Weibull distribution based on progressively type-I interval censored sample. Journal of Statistical Computation and Simulation. 2009;79(2):145‒159.
- Chen DG, Lio YL. Parameter estimations for generalized exponential distribution under progressive type-I interval censoring. Computational Statistics and Data Analysis. 2010;54(6):1581‒1591.
- Green EJ, Roesch FA, Smith AFM, et al. Bayesian estimation for the three-parameter Weibull distribution with tree diameter data. Biometrica. 1994;50(1):254‒269.
- Gupta A, Mukherjee B, Upadhyay SK. Weibull extension model: A Bayes study using Markov Chain Monte Carlo simulation. Reliability Engineering & System Safety. 2008;93(10):1434‒1443.
- Lin YJ, Lio YL. Bayesian inference under progressive type-I interval censoring. Journal of Applied Statistics. 2012;39(8):1811‒1824.
- Wu SJ, Chang CT. Parameter Estimations Based on Exponential Progressive Type II Censored with Binomial Removals. International Journal of Information and Management Sciences. 2002;13(3):37‒46.
- Kemp CD, Kemp W. Rapid generation of frequency tables. Journal of the Royal Statistical Society. 1987;36(3):277‒282.
- Efron B. The bootstrap and other resampling plans. CBMS-NSF Regional Conference Series in Applied Mathematics. SIAM, Philadelphia, USA.1982:38.
- Balakrishnan N, Sandhu RA. A simple simulational algorithm for generating progressive type-II censored samples. The American Statistician. 1995;49(2):229‒230.
- Gilks WR, Richardson S, Spiegelhalter DJ. Markov chain Monte Carlo in Practices, Chapman and Hall, London. 1996.
- Metropolis N, Rosenbluth AW, Rosenbluth MN, et al. Equations of state calculations by fast computing machine. Journal of Chemical Physics. 1953;21:1087‒1091.
- Hastings WK. Monte carlo sampling methods using markov chains and their applications. Biometrika.1970;57(1):97‒109.

©2016 Afify. This is an open access article distributed under the terms of the,
which
permits unrestricted use, distribution, and build upon your work non-commercially.


